Abstract
PROCOGNATE is a database of protein cognate ligands for the domains in enzyme structures as described by CATH, SCOP and Pfam, and is available as an interactive website or a flat file. This article gives an overview of the database and its generation and presents a new website front end, as well as recent increased coverage in our dataset via inclusion of Pfam domains. We also describe navigation of the website and its features. The current version (1.3) of PROCOGNATE covers 4123, 4536, 5876 structures and 377, 326, 695 superfamilies/families in CATH, SCOP and Pfam, respectively. PROCOGNATE can be accessed at: http://www.ebi.ac.uk/thornton-srv/databases/procognate/
INTRODUCTION
Frequently when enzyme structures are determined in vitro by X-ray crystallography or NMR, the resulting structures do not incorporate the natural substrate or product of an enzyme. Instead these ligands are often inhibitors or substrate analogues. The aim of this database is to first assign the binding of particular ligands to the evolutionary units, domains of the CATH (1), SCOP (2) and Pfam (3) databases (as observed in the experiment), and, second to make sure that the actual substrate from the enzyme's known reactions in vivo are assigned where possible. Thus, the range of actual ligands bound by a superfamily or family can be investigated. By cognate ligand, we mean one which would be found listed for that enzyme's Enzyme Commission (EC) number. We achieve this by combining data from the worldwide Protein Data Bank (wwPDB) (4) as provided in the Macromolecular Structure Database (MSD) (5), the ENZYME (6) enzyme nomenclature database and the KEGG (7) pathway database. A full description of the methodology and findings from the database can be found in Bashton et al. (8). Here we present an expanded coverage of our original dataset, notably by the addition of Pfam domain definitions and the development of a website front end.
Various other websites or databases offer some but not all of the features of PROCOGNATE. These include PDBLIG (9), BIND (10), PDBsum (11), MSDsite (12), Relibase (13) and Ligand Depot (14) but none combine information on cognate ligands and domain assignments.
Thus our database offers a unique resource in offering cognate-ligand information for domains of CATH, SCOP and Pfam and for facilitating the investigation of the evolutionary unit of proteins, domains, in relation to their molecular recognition roles.
Our database provides a list of validated cognate ligands for domains and protein structures, avoiding the problem of using data directly from the PDB where many inhibitors or substrate analogues will be present. This ‘validated’ data with corrected ligands is essential for the investigation of domain evolution and the prediction of protein function. We hope to use our data for the prediction of potential ligands bound by proteins of unknown function but known domain composition. Additionally, the database will be useful for the generation of test sets for benchmarking, programs, or methods that predict the binding of cognate ligands to proteins.
DATABASE GENERATION
This procedure involves two steps; first, we assign the binding of particular ligands to particular domains; second, we compare the chemical similarity of the PDB ligands to ligands in KEGG in order to assign cognate ligands. Database generation is automated via a series of scripts; no manual assignment is required.
Domain-ligand assignment
Binding sites may be located on different chains or even discontinuous segments of sequence. Some ligands may be bound by more than one domain, either proportionally in a shared manner, or disproportionately with the vast majority of contacts coming from one domain only. Therefore in order to produce the cognate-ligand mapping, we first assigned the binding of the PDB ligands to specific domains in protein structures.
We retrieve the total number of contacts made to any one ligand by the whole structural assembly and each domain of CATH, SCOP and Pfam in each chain from the MSD. The contact data to each ligand is retrieved from the MSD per residue level. The MSD contains contact data for the following types of bonds: hydrogen bonds, van der Waals interactions, ionic and covalent bonds, aromatic ring interactions and in absence of another type of interaction, a generic 4 Å interaction. Further details of definition of these types of bonds and interactions in the MSD can be found in Golovin et al. (12). If any one domain has greater than, or equal to, 75% of the total contacts to a particular ligand, then the binding of that ligand is assigned to that domain, and the mode of binding is recorded as ‘non-shared’. If no one domain has 75% or more of the contacts, then all contacting domains are recorded as binding the ligand and the mode of binding is recorded as ‘shared’.
Cognate-ligand assignment
All ligands in a PDB entry for a structure are compared using 2D graph matching to all compounds known to be substrates, products or cofactors for that enzyme, using data from the ENZYME and KEGG databases, and the most appropriate (i.e. chemically similar) cognate ligands are then matched up with the PDB ligands present in the PDB structure. We used 2D graph matching [using the Chemistry Development Kit libraries (15)] to compare the chemical structures of the PDB ligands and those from KEGG. We use the Tanimoto score to assess the similarity of the ligands:
where Nsub is the number of atoms in the maximum common substructure, NA is the number of atoms of molecule A and NB the number of atoms in molecule B.
In order to qualify as ‘cognate-like’, a PDB ligand needs to have a Tanimoto score of >0.5. We chose this cutoff as ∼99% of all random graph-matching scores are equal to or less than 0.5, hence we can safely consider values higher than that as significant.
Finally, the domain-ligand mapping is cross-referenced with the cognate-ligand mapping to give a cognate ligand domain mapping whereby each domain, which binds a ligand, has an assigned potential cognate taken from the various reactions catalysed by the enzyme. The similarity score of the successfully assigned potential cognate ligands are quoted on the website adjacent to each assignment.
Coverage statistics for the various versions of PROCOGNATE are given in Table 1. Coverage (in terms of the number of PDB entries) has increased 21% for CATH and 9% for SCOP since the first release of our database (8) and Pfam assignments are included for the first time in this release. The dataset is smaller than the total number of structures present in the PDB because entries need to be present as ligand-binding complexes, the proteins need to be present in CATH or SCOP, or be detectable by Pfam HMMs, and they need to have an EC number—which is also present in KEGG. Finally, the PDB ligands must be sufficiently similar to those in the KEGG reaction(s) for that structure to get an assigned cognate ligand.
Table 1.
Coverage for the various releases of PROCOGNATE. Pfam domains have only been in the dataset since version 1.3
| Version 1.3 | CATH | SCOP | Pfam |
|---|---|---|---|
| PDB entries | 4123 (21% ↑) | 4536 (9% ↑) | 5876 |
| Superfamilies/ Families | 377 | 326 | 695 |
| EC numbers | 635 | 743 | 842 |
| PDB ligands | 18731 | 20285 | 25087 |
WEBSITE: FEATURES AND NAVIGATION
The website is a live Perl-CGI generated website rendering pages dynamically based on user queries to the MySQL backend. The website can be queried at the top level by a variety of different categories; these are listed in Table 2 along with example searches to use.
Table 2.
Search categories available from the main page, examples are also provided along with description of the results of such a search
| Search category | Example string | Comments |
|---|---|---|
| PDB code | 9ldt | Leads to per PDB page view with table of domains and bound PDB ligands. For each PDB ligand, possible cognates are given along with similarity scores to the PDB ligand. |
| CATH or SCOP superfamily or Pfam family | 30.40.50.720/ c.2.1/ PF00056 | Searches with a CATH or SCOP superfamily giving families, cognate ligands, EC numbers, KEGG reactions, at family level. It also lists individual structures. |
| EC number | 1.1.1.27 | These searches return superfamilies/families and structures. |
| KEGG reaction id | R00703 | |
| KEGG compound id | C00002 | |
| PDB HET code | NAD | |
| PDB ligand name | glucose | |
| Cognate ligand name | glucose | |
| Structure title | glucose | |
| UniProt ID (primary or secondary) | P00339 or LDHA_PIG | Lists structures and chains that match that UniProt ID. |
Per PDB entry page
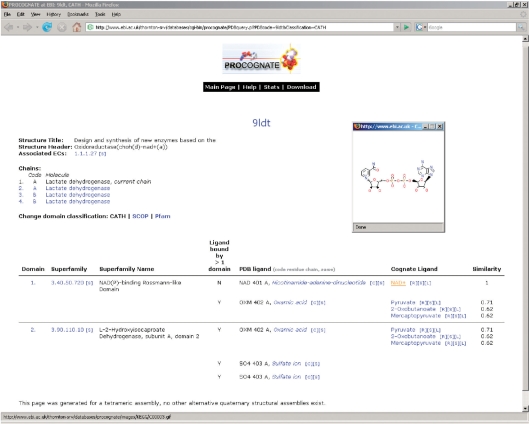
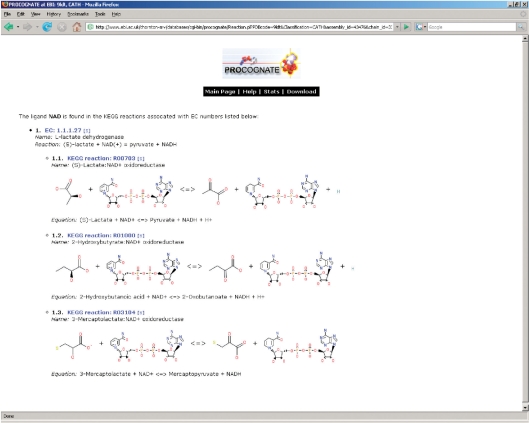
Searching with a PDB code gives a per PDB entry page overview of the domains, PDB ligands bound and assigned cognate alternatives. This page for each structure is the endpoint reached by navigating through the other search options described subsequently. Figure 1 shows an example page. This page shows the structure title, header and associated EC numbers, and chains in this assembly. A table in the centre of the page lists each domain on the currently selected chain in N- to C-terminal order. For each domain a list of bound PDB ligands, along with the mode of binding (shared, non-shared) is given in adjacent columns. Adjacent to each bound PDB ligand is a list of assigned potential cognate ligands along with a similarity score to the PDB ligand. From this page following the link for each PDB or cognate ligand will display a 2D representation of each ligand. Following the link for the domain superfamily/family identifier will redirect the browser to the relevant page in CATH, SCOP and Pfam. Additionally in the case of CATH and SCOP, the exact domain in the database can be viewed by following the link on the domain number in the first column. From this page several other functions of the website can be accessed; domains, EC number and ligands all have a search link adjacent to them, ‘[S]’ will query the database for them, the link ‘[C]’ will give a list of contacting residues to each PDB ligand and ‘[R]’ will show reactions, including diagrams for each assigned potential cognate ligand. A screen shot of the reaction page is shown in Figure 2. Links to KEGG and DrugBank (16) are also provided for each cognate ligand under ‘[L]’.
Figure 1.
Main per PDB view page for structure 9ldt. The page shows two domains, each of which binds various PDB ligands, which in turn have assigned cognate ligands. The cognate ligand NAD+ has been clicked which brings up its 2D structure in a separate window.
Figure 2.
Reaction page for NAD+ of 9ldt. Here the various EC numbers and associated KEGG reactions are shown for 9ldt, where NAD is used.
Superfamily and family searches
Searching with a SCOP or CATH superfamily will list all families in that superfamily, and in addition all cognate ligands, EC numbers and KEGG reactions associated with that superfamily. Following the link for a family will re-launch the search but at the family (rather than superfamily) level and also bring up individual structures. Searching with Pfam takes place at the family level as no subfamilies are contained within a Pfam family.
Ligand, reaction and other searches
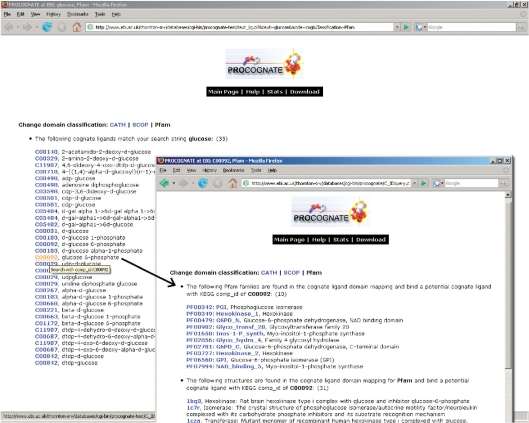
Conversely searching with a cognate or PDB ligand, EC number or KEGG reaction id will list all superfamilies/families which bind that ligand/carry out that reaction for the selected domain definition, along with all structures which bind or carry out the ligand or reaction, respectively. These searches can be restricted to a particular CATH or SCOP superfamily or a Pfam family by following the link in the results page for one of the superfamilies/families listed that bind or carry out the specified ligand or reaction. Additionally in the case of CATH and SCOP, once a search is restricted to a specific superfamily it can be further restricted to a specific family. The same functionality is available when searching with the free text name of a PDB or cognate ligand or structure title. A PDB or cognate ligand name can also be used to initiate a search. This will retrieve a list of ligand identifiers whose names contain the search string. Selecting one of these the search will continue in the same way as those described above. Figure 3 shows an example of searching with a cognate ligand name. Finally searching with a UniProt (17), primary or secondary id will give a list of PDB codes and chains that correspond to that identifier. Selecting one of these will give the per PDB code page for that entry with the chain corresponding to the given UniProt ID pre-selected.
Figure 3.
The results of searching for cognate ligand name glucose. The search first returns a list of cognate ligands with the text glucose in their name. Clicking on one of these then searches with that particular ligand—this is shown in the second screen shot on the right.
FLAT FILE DOWNLOAD
Our database is freely available; the tab delimited flat file for all versions of PROCOGNATE for each different domain definition can be downloaded from http://www.ebi.ac.uk/thornton-srv/databases/procognate/download.html.
FUTURE DEVELOPMENTS
Currently the website focuses on providing interactive access and facilitating querying the database backend providing cognate-ligand assignments for structures of enzymes in the PDB. We aim to expand the functionality of the website to offer a prediction of ligand binding for both user-submitted sequences and structures based on similarity to the known domains in our database and their ligand-binding profiles.
ACKNOWLEDGEMENTS
M.B. was supported by NIH grant (GM62414), US DOE under contract (W-31-109-ENG38). I.N. gratefully acknowledges financial support from the Medical Research Council in the form of a Training Fellowship in Bioinformatics for the period 2001 to 2005. Funding to pay the Open Access publication charge was provided by NIH grant (GM62414), US DOE under contract (W-31-109-ENG38).
Conflict of interest statement. None declared.
REFERENCES
- 1.Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH – a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 2.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 3.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 5.Velankar S, McNeil P, Mittard-Runte V, Suarez A, Barrell D, Apweiler R, Henrick K. E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res. 2005;33:D262–D265. doi: 10.1093/nar/gki058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bashton M, Nobeli I, Thornton JM. Cognate ligand domain mapping for enzymes. J. Mol. Biol. 2006;364:836–852. doi: 10.1016/j.jmb.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 9.Chalk AJ, Worth CL, Overington JP, Chan AW. PDBLIG: classification of small molecular protein binding in the Protein Data Bank. J. Med. Chem. 2004;47:3807–3816. doi: 10.1021/jm040804f. [DOI] [PubMed] [Google Scholar]
- 10.Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K, Betel D, Bobechko B, Boutilier K, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005;33:D266–D268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golovin A, Dimitropoulos D, Oldfield T, Rachedi A, Henrick K. MSDsite: a database search and retrieval system for the analysis and viewing of bound ligands and active sites. Proteins. 2005;58:190–199. doi: 10.1002/prot.20288. [DOI] [PubMed] [Google Scholar]
- 13.Hendlich M. Databases for protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 1998;54:1178–1182. doi: 10.1107/s0907444998007124. [DOI] [PubMed] [Google Scholar]
- 14.Feng Z, Chen L, Maddula H, Akcan O, Oughtred R, Berman HM, Westbrook J. Ligand Depot: a data warehouse for ligands bound to macromolecules. Bioinformatics. 2004;20:2153–2155. doi: 10.1093/bioinformatics/bth214. [DOI] [PubMed] [Google Scholar]
- 15.Steinbeck C, Han Y, Kuhn S, Horlacher O, Luttmann E, Willighagen E. The Chemistry Development Kit (CDK): an open-source Java library for Chemo- and Bioinformatics. J. Chem. Inf. Comput. Sci. 2003;43:493–500. doi: 10.1021/ci025584y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]