Abstract
The identification of orthologous genes forms the basis for most comparative genomics studies. Existing approaches either lack functional annotation of the identified orthologous groups, hampering the interpretation of subsequent results, or are manually annotated and thus lag behind the rapid sequencing of new genomes. Here we present the eggNOG database (‘evolutionary genealogy of genes: Non-supervised Orthologous Groups’), which contains orthologous groups constructed from Smith–Waterman alignments through identification of reciprocal best matches and triangular linkage clustering. Applying this procedure to 312 bacterial, 26 archaeal and 35 eukaryotic genomes yielded 43 582 course-grained orthologous groups of which 9724 are extended versions of those from the original COG/KOG database. We also constructed more fine-grained groups for selected subsets of organisms, such as the 19 914 mammalian orthologous groups. We automatically annotated our non-supervised orthologous groups with functional descriptions, which were derived by identifying common denominators for the genes based on their individual textual descriptions, annotated functional categories, and predicted protein domains. The orthologous groups in eggNOG contain 1 241 751 genes and provide at least a broad functional description for 77% of them. Users can query the resource for individual genes via a web interface or download the complete set of orthologous groups at http://eggnog.embl.de.
INTRODUCTION
The vast majority of the functionally annotated genes in genomes or metagenomes are derived by comparative analysis and inference from existing functional knowledge via homology. With the sequencing of entire genomes, it became possible to increase the resolution of the functional transfer by distinguishing between orthologs and paralogs, that is gene pairs that trace back to speciation and gene duplication events, respectively (1). These concepts have since been extended and refined to include orthologous groups (2), in-paralogs and out-paralogs (3,4), but the identification and classification of homologous genes remains very difficult. In contrast to the definition of orthology, the classification of genes into orthologous groups is always with respect to a taxonomic position: two paralogous genes from human and mouse may be orthologs of the same gene in fruit fly and will belong to either the same or different orthologous groups depending on whether these are defined with respect to the last common ancestor of metazoans or mammals. This is further complicated by evolutionary processes such as gene fusion and domain shuffling, due to which each domain of a multi-domain protein is not guaranteed to have evolved through the same series of speciation and duplication events. Finally, because we do not know how each gene evolved, one in practice always relies on operational definitions rather than the evolutionary definitions given above.
Numerous methods have been developed to derive orthologs and orthologous groups, ranging from the simple reciprocal-best-hit approach, via InParanoid (5), MultiParanoid (6), identification best-hit triangles (2,7,8) and clustering-based approaches (9), to tree-based methods (10–13). By contrast, there has been only one major effort to provide functionally annotated orthologous groups, namely the COG/KOG database (2,8), but it lacks phylogenetic resolution and is not regularly updated due to the manual labor required. There is thus a need for a hierarchical system of orthology classification with function annotation.
Here, we provide such a system, eggNOG, which (1) can be updated without the requirement for manual curation, (2) covers more genes and genomes than existing databases, (3) contains a hierarchy of orthologous groups to balance phylogenetic coverage and resolution and (4) provides automatic function annotation of similar quality to that obtained through manual inspection.
CONSTRUCTION OF HIERARCHICAL ORTHOLOGOUS GROUPS
We assemble proteins into orthologous groups using an automated procedure similar to the original COG/KOG approach (2,8). When constructing coarse-grained orthologous groups across all three domains of life or for all eukaryotes, we first assign the proteins encoded by the genomes in eggNOG to the respective COGs or KOGs based on best hits to the manually assigned sequences in the COG/KOG database. In case of multiple hits to the same part of the sequence, only the best hit was considered. The many proteins that cannot be assigned to existing COGs or KOGs are subsequently assembled into non-supervised orthologous groups using the procedure described below. When constructing more fine-grained orthologous groups, this initial step is skipped.
Briefly, we first compute all-against-all Smith–Waterman similarities among all proteins in eggNOG. We then group recently duplicated sequences into in-paralogous groups, which are subsequently treated as single units to ensure that they will be assigned to the same orthologous groups. To form the in-paralogous groups, we first assemble highly related genomes into clades, usually encompassing all sequenced strains of a particular species in a single clade, but also other close pairs such as human and chimpanzee. In these clades, we join into in-paralogous groups all proteins that are more similar to each other (within the clade), than to any other protein outside the clade. For this, there is no fixed cutoff in similarity, but instead we start with a stringent similarity cutoff and relax it a step-wise fashion until all in-paralogous proteins are joined, requiring that all members of a group must align to each other with at least 20 residues.
After grouping in-paralogous proteins, we start assigning orthology between proteins, by joining triangles of reciprocal best hits involving three different species (here, in-paralogous groups are represented by their best-matching member). Again, we start with a stringent similarity cutoff and relax it to identify groups of proteins that all align to each other by at least 20 residues. This procedure occasionally causes an orthologous group to be split in two; such cases are identified by an abundance of reciprocal best hits between groups, which are then joined. Next, we relax the triangle criterion and allow remaining unassigned proteins to join a group by simple bidirectional best hits. Finally, we automatically identify gene fusion events by searching for proteins that bridge otherwise unrelated orthologous groups. In these cases, the different parts of the fusion protein are assigned to their respective orthologous groups. This step is a distinguishing feature of our approach and is crucial for the analysis of eukaryotic multi-domain proteins, as these would otherwise cause unrelated orthologous groups to be fused.
To construct a hierarchy of orthologous groups, the procedure described above was applied to several subsets of organisms. To make a set of course-grained orthologous groups across all three domains of life, we constructed non-supervised orthologous groups (NOGs) from the genes that could not be mapped to a COG or KOG. Focusing on eukaryotic genes, we constructed more fine-grained eukaryotic NOGs (euNOGs) from the genes that could not be mapped to a KOG. Finally, we build sets of NOGs of increasing resolution for five eukaryotic clades, namely fungi (fuNOGs), metazoans (meNOGs), insects (inNOGs), vertebrates (veNOGs) and mammals (maNOGs).
AUTOMATIC ANNOTATION OF PROTEIN FUNCTION
An important feature of eggNOG is that it provides functional annotations for the orthologous groups. These annotations are produced by a pipeline, which summarizes the available functional information on the proteins in each cluster: (1) the textual annotation for these proteins, (2) their annotated Gene Ontology (GO) terms (14), (3) their membership to KEGG pathways (15) and (4) the presence of protein domains from SMART (16) and Pfam (17). As the textual descriptions allow for the most fine-grained annotation of protein function, we first use Ukkonen's; algorithm (18) to identify the longest common subsequence (LCS) between the description lines of any two proteins within a cluster. We then score each LCS based on the number of protein descriptions matched within the cluster, the number of occurrences of each word of the LCS in these descriptions, and the presence of words such as ‘hypothetical’, ‘putative’ or ‘unknown’. These scores are finally normalized against a score distribution based on randomized clusters of the same size, and the highest scoring LCS is chosen, provided that it scores above a threshold.
For each orthologous group, our pipeline also searches for overrepresented GO terms, KEGG pathways or protein domains. To find terms that are sufficiently specific and at the same time are likely to describe the entire orthologous group, we devised a scoring function that takes into account term frequency within the group, background frequency, and the ratio of the two (i.e. the fold overrepresentation). In case no satisfactory LCS was found, a description line is constructed based on the highest scoring GO term or KEGG pathway. As a single domain may not properly reflect the function of a complete protein, description lines are constructed based on overrepresented domains only if all other options have been exhausted.
QUALITY ASSESSMENT AND SUMMARY STATISTICS
To assess the quality of the function annotations provided by our automated pipeline, we manually checked a random sample of 100 NOGs and 100 euNOGs and classified their annotations into three categories: 87.5% were correct (i.e. they describe a function that the proteins have in common), 12.5% were uninformative (i.e. they do not describe a function) and, due to our stringent rule set, no wrong functions were assigned. Uninformative annotations of orthologous groups are in many cases due to a lack of functional knowledge on the corresponding proteins.
Our function annotation pipeline enables us to provide description lines for 6583 of the 33 858 (19%) coarse-grained NOGs. Combined with the 9724 COGs and KOGs, this yields 43 582 global orthologous groups of which 14 356 (33%) have an annotated function. In addition, eggNOG contains 94 240 more fine-grained orthologous groups of which 55 753 (59%) could be functionally annotated. This enables us to assign 1 241 751 of 1 513 782 genes (82% of the genes in the analyzed genomes) to an orthologous group and to provide at least a broad functional description of 951 918 of them (77% of the genes that could be assigned to an orthologous group). The corresponding numbers for each set of orthologous groups as well as for each individual genome are summarized in Figure 1.
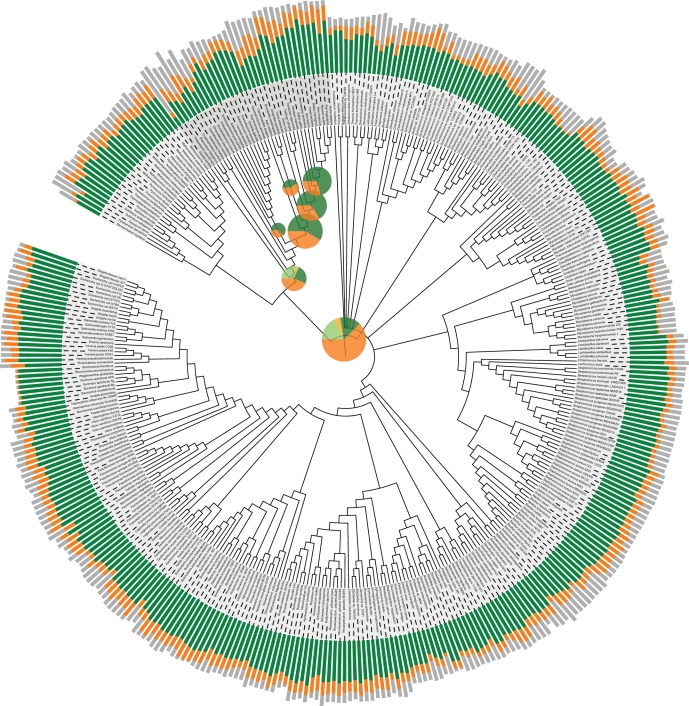
Figure 1.
Statistics on the content of the eggNOG database. The eggNOG assignments for 373 complete genomes [19] were mapped onto the tree of life. The stacked bar charts outside the tree show the proportion of genes from each genome that can be assigned to a functionally annotated orthologous group (green), to an unannotated orthologous group (orange) or to no orthologous group (grey). The length of each bar is proportional to the logarithm of the number of genes in the respective genome. The pie charts inside the tree show the fractions of orthologous groups at each level in the hierarchy that could be annotated with a function description (green for NOGs, light green for extended COGs and KOGs) and that could not be functionally annotated (orange for NOGs, light orange for extended COGs and KOGs). The areas of the pie charts are proportional to the number of orthologous groups at the phylogenetic level in question. This figure was made using iTOL [20].
Using eggNOG
The eggNOG resource is accessible via a web interface at http://eggnog.embl.de. The main page allows the user to input the names of one or more genes or orthologous groups and to optionally select the organism of interest. Alternatively, the user can choose to upload a set of protein sequences to be searched against the full-length sequences in eggNOG. In case of ambiguous names or query sequences with multiple hits, the user is prompted to disambiguate the input.
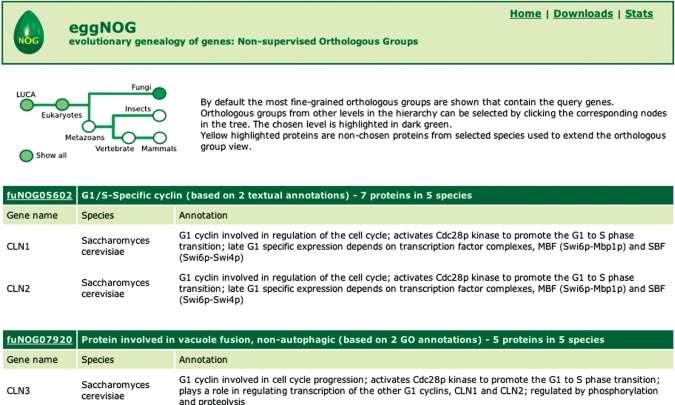
Figure 2 shows the result of a query for the three G1-type cyclins in budding yeast, which belong to two distinct fungal orthologous groups. Function descriptions are displayed for both the orthologous groups and for the individual genes. The web interface enables the user to view the complete set of genes that belong to each orthologous group and provides external links to additional information on the protein products.
Figure 2.
Screenshot of the main results page. The eggNOG database was queried for the three G1-type cyclins in budding yeast, namely Cln1–Cln3. These have been correctly assigned to two fungal orthologous groups. The navigation tree at the top of the page allows the user to change the view to more coarse-grained orthologous groups, for example the eukaryotic orthologous groups in which these cyclins are all grouped together.
By default, eggNOG shows the most fine-grained orthologous groups that are possible given the input: just like entering a set of genes from budding yeast results in fungal orthologous groups being shown, a set of human genes will yield mammalian orthologous groups, whereas a combination of human and fruit fly genes will yield metazoan orthologous groups. A navigation tree at the top of the page (Figure 2) allows the user to select more coarse-grained orthologous groups if desired; for example, selecting ‘eukaryotes’ reveals that the three budding yeast cyclins all belong to the same eukaryotic orthologous group. This key feature enables the user to choose the balance between phylogenetic coverage and resolution within our hierarchy of orthologous groups.
Whereas the web interface is convenient for small-scale studies, users interested in genome-wide analyses will be better served by downloading the complete content of the underlying relational database. For this reason, the orthologous groups, functional annotations and protein sequences are all available from the eggNOG download page under the Creative Commons Attribution 3.0 License.
ACKNOWLEDGEMENTS
The authors thank Eugene Koonin for comments on the manuscript. This work was supported by Bundesministerium für Bildung und Forschung (Nationales Genomforschungsnetz grant 01GR0454) as well as through the GeneFun Specific Targeted Research Project, contract number LSHG-CT-2004-503567, and through the BioSapiens Network of Excellence, contract number LSHG-CT-2003-503265, both funded by the European Commission FP6 Programme. Funding to pay the Open Acces publication charges this article was provided by the European Molecular Biology Laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fitch WM. Distinguishing homologous from analogous proteins. J. Biol. Chem. 1970;19:99–113. [PubMed] [Google Scholar]
- 2.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 3.Sonnhammer EL, Koonin EV. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- 4.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 5.O'Brien KP, Remm M, Sonnhammer ELL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexeyenko A, Tamas I, Liu G, Sonnhammer ELL. Automatic clustering of orthologs and inparalogs shared by multiple proteomes. Bioinformatics. 2006;14:e9–e15. doi: 10.1093/bioinformatics/btl213. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Sultana R, Pertea G, Cho J, Karamycheva S, Tsai J, Parvizi B, Cheung F, et al. Cross-referencing eukaryotic genomes: TIGR orthologous gene assignments (TOGA) Genome Res. 2002;12:493–502. doi: 10.1101/gr.212002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov S, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of orthologous groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Coghlan A, Ruan J, Coin LJ, Hériché J-K, Osmotherly L, Li R, Liu T, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Heijden RTJM, Snel B, van Noort V, Huynen MA. Orthology prediction at scalable resolution by phylogenetic tree analysis. BMC Bioinformatics. 2007;8:83. doi: 10.1186/1471-2105-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wapinski I, Pfeffer A, Friedman N, Regev A. Automatic genome-wide reconstruction of phylogenetic trees. Bioinformatics. 2007;23:i549–i558. doi: 10.1093/bioinformatics/btm193. [DOI] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight S, et al. Gene Ontology: tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letunic I, Copley RR, Schmidt S, Ciccarelli F, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ukkonen E. On-line construction of suffix trees. Algorithmica. 1995;14:249–260. [Google Scholar]
- 19.von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Krüger B, Snel B, Bork P. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]