Abstract
Mitome is a specialized mitochondrial genome database designed for easy comparative analysis of various features of metazoan mitochondrial genomes such as base frequency, A+T skew, codon usage and gene arrangement pattern. A particular function of the database is the automatic reconstruction of phylogenetic relationships among metazoans selected by a user from a taxonomic tree menu based on nucleotide sequences, amino acid sequences or gene arrangement patterns. Mitome also enables us (i) to easily find the taxonomic positions of organisms of which complete mitochondrial genome sequences are publicly available; (ii) to acquire various metazoan mitochondrial genome characteristics through a graphical genome browser; (iii) to search for homology patterns in mitochondrial gene arrangements; (iv) to download nucleotide or amino acid sequences not only of an entire mitochondrial genome but also of each component; and (v) to find interesting references easily through links with PubMed. In order to provide users with a dynamic, responsive, interactive and faster web database, Mitome is constructed using two recently highlighted techniques, Ajax (Asynchronous JavaScript and XML) and Web Services. Mitome has the potential to become very useful in the fields of molecular phylogenetics and evolution and comparative organelle genomics. The database is available at: http://www.mitome.info.
INTRODUCTION
Metazoan mitochondrial genomes have special features such as maternal inheritance, rapid evolutionary rate, lack of introns, lack of recombination events and haploidy (1,2). In consequence, various types of metazoan mitochondrial genome information—genome organization, gene order and orientation, AT skewness, codon usage, base frequency, base ratio, codon usage pattern, secondary structures of tRNA and rRNA, etc.—are providing important clues for solving enigmas in the fields of molecular evolution, population genetics and molecular phylogenetics (3–5). In particular, comparative analyses of mitochondrial gene arrangements have great potential for elucidating metazoan evolutionary relationships because almost all metazoan species have the same set of well-characterized gene components and the order of this set rarely changes (6).
The increasing importance of mitochondrial genomes has led to the development of various mitochondrial genome-related databases: Gobase (7), OGRe (8), AMiGA (9), the Joint Genome Institute Organelle Database (http://evogen.jgi.doe.gov/top_level/organelles.html), MamMiBase (10) and the NCBI Organelle Resources database (11). These are useful for retrieving basic information about complete metazoan mitochondrial genomes and comparing their gene arrangement patterns, and have also played key roles in developing search engines that are more refined than primary public repositories. However, as the number of completely sequenced metazoan mitochondrial genomes is rapidly increasing (over 1000 species of 19 metazoan phyla) and various modes of data analysis are now required, there is a need for a new multi-functional mitochondrial genome database implementing dynamic and more complex web-based applications. We have therefore attempted to develop a powerful, user-friendly web-based database with multiple functions that can analyze a variety of mitochondrial genome characteristics simultaneously, in addition to the basic functions of the previously constructed databases.
A web-based application has many advantages: it can reach many users, there is no need to install it, and it is easier to update and manage from desktops. However, such applications are less dynamic and interactive than traditional desktop applications. In addition, a web-based application handling large volumes of information imposes irritatingly long loading and analyzing times on the users. Two techniques to overcome these disadvantages have recently been highlighted: Ajax (Asynchronous JavaScript and XML) and Web Services. These allow users to be provided with dynamic and responsive web-based applications. Ajax, based on JavaScript and HTTP, uses JavaScript to send and receive data between a web browser and a web server (http://www.openajax.org). Web services are software systems designed for machine-to-machine communication over a network such as the Internet and for executing a remote program hosting the requested services (http://www.w3.org/TR/2007/REC-wsdl20-primer-20070626).
We have developed a dynamic and interactive multifunctional web-based database, Mitome, using Ajax and Web Services. Mitome will help researchers to elucidate metazoan evolutionary relationships inferred from mitochondrial genome information and to carry out comparative analyses of mitochondrial genome sequences and arrangement patterns, as well as extracting useful basic information such as AT skewness, codon usage pattern, base frequency, etc. from the mitochondrial genomes selected by users. Mitome is a user-friendly database with a web-based application, which, dealing with many data at a time, is better, faster and more interactive than traditional web-based applications.
CONTENTS OF MITOME
Complete sequences of mitochondrial genomes from 1056 metazoan species (in August 2007) are listed in the taxonomic tree (Table 1). The users can easily select any interesting taxa from the set and conduct various further analyses using the implemented applications. The set is automatically updated when new data are uploaded in the primary public databases such as NCBI, EMBL and DDBJ. Details of the main functions and features of Mitome are as follows.
Table 1.
Contents of the Mitome at the time of submission (August 2007)
| Metazoan phyla | Number of genomes |
|---|---|
| Acanthocephala | 1 |
| Annelida | 4 |
| Arthropoda | 132 |
| Brachiopoda | 3 |
| Bryozoa | 1 |
| Chaetognatha | 2 |
| Chordata | 783 |
| Cnidaria | 24 |
| Echinodermata | 16 |
| Echiura | 1 |
| Hemichordata | 2 |
| Mollusca | 34 |
| Nematoda | 21 |
| Onychophora | 1 |
| Placozoa | 4 |
| Platyhelminthes | 20 |
| Porifera | 5 |
| Priapulida | 1 |
| Xenoturbellida | 1 |
| Mitome Total | 1056 |
Sequence analyses: Comparative analyses of nucleotide and/or amino acid sequences
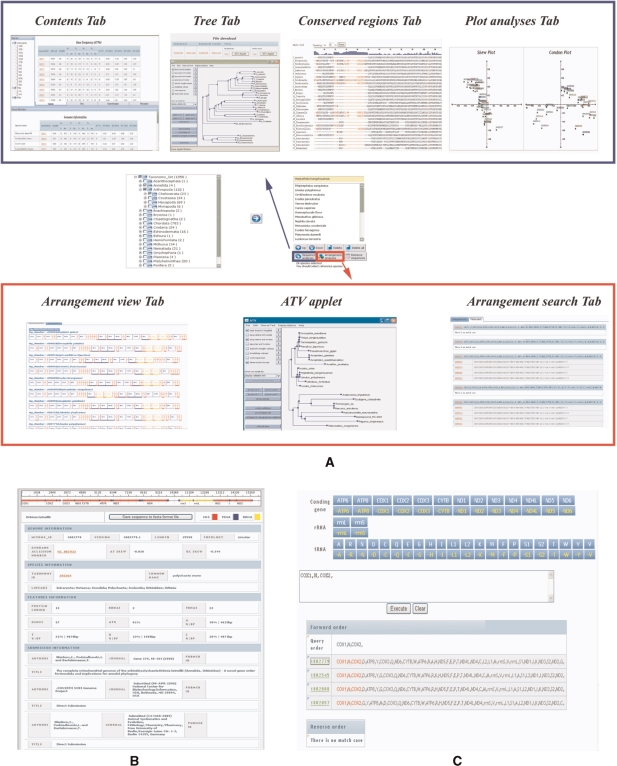
When users conduct comparative analyses of the nucleotide and/or amino acid sequences of each mitochondrial gene, they can choose four different tabs under Sequence analyses—Contents, Trees, Conserved regions and Plot analyses (Figure 1A, see the blue box).
Contents. The Contents tab includes two different tables showing base frequency and codon analysis. The base frequency table provides basic statistical information about total gene length, base composition, percentage AT content, AT skewness, GC skewness, AT ratio and GC ratio. The codon analysis table provides a codon usage table and values meaningful for inferring codon usage patterns.
Trees. This tab is a particular feature of Mitome. Nucleotide and amino acid sequences of each gene in the organisms selected by users are multi-aligned using ClustalW-MPI (12) to reduce computer calculation time. The application of ClustalW-MPI to Mitome makes it possible to perform analyses of massive amounts of sequence data online relatively quickly. The neighbor-joining tree inferred from the constructed sequence alignment is visualized via ATV software (13). It is possible to download not only the resultant alignment files of amino acid and/or nucleotide sequences but also the tree files for the selected organisms.
Conserved regions. This tab shows the percentage identities between certain interesting sequences and a reference sequence selected by the user. The reference sequence is always shown at the top of the sequence alignment. It is helpful for finding conserved regions and exploring stable primer sites. Amino acid or nucleotide sequences with over 70% identities to the default option are shown in red. Users can change the lower percentage identity limit.
Plot analyses. Both skew and codon plots are displayed in this tab. The skew plot shows AT and GC skewnesses along the forward and reverse strands. The codon plot depicts the results of correspondence analyses performed by a CodonW program (14). The spot distribution pattern provides general information with respect to the base deviation index and codon usage preference.
Figure 1.
(A) Comparative analyses. Red and blue boxes show, respectively, the four tabs in the Sequence analyses part and the two tabs in the Arrangements analyses part. (B) Genome browser. This provides detailed information related to genome, species and submission/reference by clicking a particular organism on the taxonomy tree menu. (C) Pattern search. This is useful for finding organisms with mitochondrial gene orders specified by users.
Arrangement analyses: Comparative analyses of mitochondrial gene arrangements
Mitochondrial gene arrangements can be analyzed using two tabs—Arrangement view and Pattern match—in the Arrangement analyses part (Figure 1A, see the red box). Through Arrangement view, users can compare gene arrangements corresponding to the selected metazoans in a browser. A particular function of Mitome is to reconstruct a phylogenetic tree with a dissimilarity matrix among gene arrangement data. The resultant tree is also visualized using ATV software. From this tab, users can download all the information and analysis results mentioned above: graphic views of gene arrangements, dissimilarity matrices and phylogenetic trees.
Pattern match enables users to search organisms with arrangement patterns identical to those of the organisms they have selected on the taxonomy tree in Mitome.
Retrieve FASTA files
When using the Comparative Analysis entry, the Retrieve sequences button on the selected taxa window allows users to be able to download each sequence of 37 genes coding for 13 proteins, 2 rRNAs and 22 tRNAs. Mitome also generates FASTA files for all of them, which makes it feasible to conduct further studies such as phylogenetic and evolutionary analyses.
Genome browser
The genome browser of Mitome shows annotation features of whole mitochondrial genomes by clicking a particular organism on the taxonomy tree menu (Figure 1B). The web-based genome browser was implemented using JavaServer Faces (JSF) technology, which has the advantage of constructing a clearly defined architecture by separating application logic and presentation. This browser shows graphical views of mitochondrial genomes in linear forms. In the picture, protein-coding regions (CDS) are in red, tRNA-coding regions in blue and rRNA-coding regions in yellow. A good deal of further information related to genomes, species and submission/reference is described in detail on the web browser. In addition, the genome browser provides the corresponding ID numbers of GenBank, PubMed or Taxonomy, which are directly linked. Users can also download a FASTA file containing whole mitochondrial genome sequences.
Pattern search
Pattern search allows users to enter a specific gene order into the text box and search for best-matching gene arrangements through Mitome (Figure 1C). By entering a specific gene order, users can click the corresponding gene buttons in order, or write directly into the text box. In the pattern search menu, organisms with arrangement patterns identical to those selected are listed with mg_number, and matched regions are shown in red. By clicking mg_number, users can enter the corresponding genome browser.
SYSTEM AND IMPLEMENTATION
Mitome is a fast and interactive web-based database with a user-friendly interface. If users select a particular organism on the taxonomy tree menu, they can acquire all the information they need from a MySQL relational database in which all metazoan mitochondrial genome information is stored. All the information in this database was extracted from GenBank, PubMed and NCBI-Taxonomy. The stored data are automatically updated daily. Its own analyzing tools written in Java are implemented and the results of analyses performed by the tools are automatically stored in Mitome. The web interface is based on a Tomcat 5.5 server with dynamic contents generated by JSP and AJAX, with the aim of speeding up the sending and receiving of data between a web browser and a web server. To perform multiple sequence comparisons and to examine phylogentic relationships among selected organisms, all genes coding for proteins, rRNAs and tRNAs are first aligned using ClustalW-MPI. This runs through Web Services on clustered computers with 20 nodes (Intel Xeon 3.06 GHz with 2 GB RAM) to reduce the time consumed in analyzing masses of data from a number of metazoan species simultaneously. However, in constructing nucleotide sequence alignments of protein-coding genes, the corresponding proteins are first aligned with ClustalW-MPI and then the nucleotide sequences are aligned by reference to the amino acid sequence alignment using RoCDS (unpublished data) written in Java.
FUTURE DEVELOPMENTS
Analysis tools such as BLAST search, automatic annotation, RNA secondary structure prediction, a much more user-friendly submission system and so on will continue to be added to Mitome. To date, Mitome includes only those mitochondrial genome sequences obtainable from public databases (e.g. GenBank). We will implement a new, powerful submission system specialized for mitochondrial genomes. This will promote the depositing of unpublished mitochondrial genome data into Mitome, and it will help users to analyze their own data easily using the powerful web-based applications implemented in Mitome.
Chloroplast genome data have also been widely used as genetic markers for phylogenetic and evolutionary studies in plants (15,16). As more and more complete chloroplast genome data become available, the need for a web-based chloroplast genome database akin to Mitome increases. The platform of Mitome developed in the present study could make it easier to construct a web-based chloroplast genome database in the near future.
ACKNOWLEDGEMENTS
This work was supported by a Korea Science and Engineering Foundation (KOSEF) grant funded by the Korean government (Ministry of Science and Technology) (No. R01-2004-000-10930-0), a Research Initiative Program of Korea Research Institute for Bioscience & Biotechnology (KRIBB), and a Korea Institute of Environmental Science and Technology (KIEST) grant funded by the Korean government (Ministry of Environment) (No. 2007-05002-0051-0) awarded to U.W.H. Funding to pay the Open Access publication charges for this article was provided by the KIEST grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc. Natl Acad. Sci. USA. 1980;77:6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown WM, George M, Jr, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc. Natl Acad. Sci. USA. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassanin A, Leger N, Deutsch J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Syst. Biol. 2005;54:277–298. doi: 10.1080/10635150590947843. [DOI] [PubMed] [Google Scholar]
- 4.Lavrov DV, Forget L, Kelly M, Lang BF. Mitochondrial genomes of two demosponges provide insights into an early stage of animal evolution. Mol. Biol. Evol. 2005;22:1231–1239. doi: 10.1093/molbev/msi108. [DOI] [PubMed] [Google Scholar]
- 5.Ballard JW, Whitlock MC. The incomplete natural history of mitochondria. Mol. Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 6.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Brien EA, Zhang Y, Yang L, Wang E, Marie V, Lang BF, Burger G. GOBASE – a database of organelle and bacterial genome information. Nucleic Acids Res. 2006;34:D697–D699. doi: 10.1093/nar/gkj098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jameson D, Gibson AP, Hudelot C, Higgs PG. OGRe: a relational database for comparative analysis of mitochondrial genomes. Nucleic Acids Res. 2003;31:202–206. doi: 10.1093/nar/gkg077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feijao PC, Neiva LS, de Azeredo-Espin AM, Lessinger AC. AMiGA: the arthropodan mitochondrial genomes accessible database. Bioinformatics (Oxf., Engl.) 2006;22:902–903. doi: 10.1093/bioinformatics/btl021. [DOI] [PubMed] [Google Scholar]
- 10.Vasconcelos AT, Guimaraes AC, Castelletti CH, Caruso CS, Ribeiro C, Yokaichiya F, Armoa GR, Pereira Gda S, da Silva IT, et al. MamMiBase: a mitochondrial genome database for mammalian phylogenetic studies. Bioinformatics (Oxf., Engl.) 2005;21:2566–2567. doi: 10.1093/bioinformatics/bti326. [DOI] [PubMed] [Google Scholar]
- 11.Wolfsberg TG, Schafer S, Tatusov RL, Tatusov TA. Organelle genome resource at NCBI. Trends Biochem. Sci. 2001;26:199–203. doi: 10.1016/s0968-0004(00)01773-4. [DOI] [PubMed] [Google Scholar]
- 12.Li KB. ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics (Oxf., Engl.) 2003;19:1585–1586. doi: 10.1093/bioinformatics/btg192. [DOI] [PubMed] [Google Scholar]
- 13.Zmasek CM, Eddy SR. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics (Oxf., Engl.) 2001;17:383–384. doi: 10.1093/bioinformatics/17.4.383. [DOI] [PubMed] [Google Scholar]
- 14.Shil P, Dudani N, Vidyasagar PB. ISHAN: sequence homology analysis package. In silico Biol. 2006;6:373–377. [PubMed] [Google Scholar]
- 15.Clegg MT. Chloroplast gene sequences and the study of plant evolution. Proc. Natl Acad. Sci. USA. 1993;90:363–367. doi: 10.1073/pnas.90.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cattolico RA. Chloroplast biosystematics: chloroplast DNA as a molecular probe. Bio Syst. 1985;18:299–306. doi: 10.1016/0303-2647(85)90030-9. [DOI] [PubMed] [Google Scholar]