Abstract
The Stanford Tissue Microarray Database (TMAD; http://tma.stanford.edu) is a public resource for disseminating annotated tissue images and associated expression data. Stanford University pathologists, researchers and their collaborators worldwide use TMAD for designing, viewing, scoring and analyzing their tissue microarrays. The use of tissue microarrays allows hundreds of human tissue cores to be simultaneously probed by antibodies to detect protein abundance (Immunohistochemistry; IHC), or by labeled nucleic acids (in situ hybridization; ISH) to detect transcript abundance. TMAD archives multi-wavelength fluorescence and bright-field images of tissue microarrays for scoring and analysis. As of July 2007, TMAD contained 205 161 images archiving 349 distinct probes on 1488 tissue microarray slides. Of these, 31 306 images for 68 probes on 125 slides have been released to the public. To date, 12 publications have been based on these raw public data. TMAD incorporates the NCI Thesaurus ontology for searching tissues in the cancer domain. Image processing researchers can extract images and scores for training and testing classification algorithms. The production server uses the Apache HTTP Server, Oracle Database and Perl application code. Source code is available to interested researchers under a no-cost license.
INTRODUCTION
The Tissue Microarray Database (TMAD; http://tma.stanford.edu) at Stanford University is a web-based system that provides researchers with tissue microarray design tools, image scoring and annotation tools, data sharing mechanisms, an image archive, an analysis toolset and publication mechanism. Tissue microarray experiments provide in situ detection of protein, DNA and RNA targets on hundreds of tissue specimens per slide through chromogenic and fluorescence stains. Images at subcellular resolution of each specimen are taken for subsequent scoring and analysis. Each image is rich in multivariate information including cell composition and morphology as well as stain localization.
In 1987, Wan et al. (1) described a method to immunohistochemically stain many different tissues simultaneously on a single slide, the stated advantages being great economies in time, reagents, tissue specimens and antibodies. Tissue microarrays in their current form were developed by Kallioniemi and Sauter (2) for high-throughput molecular profiling of tissue specimens.
Twenty years later these advantages have proven to be true, and today the Stanford Tissue Microarray Database contains over 200 000 stained and scored tissue microarray images along with associated tissue metadata describing the tissues, associated clinical diagnosis and follow-up where available. TMAD includes tools for tissue microarray design, image and scoring import, and analysis tools via an intuitive web interface.
Several database object models (3,4) and systems (5–10) have been described for managing tissue microarray data. Goals range from metadata modeling to comprehensive management of tissue microarrays for large research groups. While there are similarities, TMAD differs by providing public access to raw tissue microarray experiment data. As part of ongoing collaborations with non-US research groups we have constructed a straightforward method to import images and metadata from collaborating institutions, eliminating sample and slide transportation between institutes and resulting complications and delays.
The Human Protein Atlas project (11,12) has published a comprehensive public access antibody-based protein atlas based on the systematic creation of protein-specific antibodies applied to tissue microarrays and used to create expression and localization profiles in 48 normal human tissues, 20 varied cancers as well as 47 cell lines. Their version 2.0 Atlas available at http://www.proteinatlas.org/ includes over 1 200 000 images corresponding to over 1500 antibodies. We believe that TMAD provides a complementary service with selected probe data across a wider variety of disease tissues along with an integrated tissue microarray toolset.
The Nordic Immunohistochemical Quality Control organization (13) publishes very detailed IHC results including thousands of images for clinically important epitopes. Their data comes from over 100 laboratories that participate in quality control studies by performing independent stains on serial sections of multiple tissue blocks that are then verified independently. Their in-depth information on antigens and protocols is available at http://www.nordiqc.org/. While TMAD includes standard clinical antibody probes, it adds many novel emerging antibody probes useful for the molecular sub-classification of cancers.
PUBLIC ACCESS
We designed TMAD to allow for the release of raw supporting data (including images) at the time of publication for all experiments held in TMAD. Researchers using TMAD observe a policy of making data publicly available through TMAD at the point of publication (or earlier) (14–20). We have implemented automated mechanisms that allow tagging the complete set of experiments associated with each new publication, resulting in nearly ‘one click’ publication of the raw data (stained images and scores assigned by pathologists) through TMAD.
As of July 2007, TMAD contained 205 161 images archiving 349 distinct probes on 1488 stained tissue microarray slides. Of these, 31 306 images for 68 probes on 125 slides have been released to the public.
By focusing on the release of data for public use, we anticipate improved collaboration among data model and database developers. Our ‘real world’ data can be used to validate both object models and eXtensible Markup Language (XML) (21) based tissue microarray data exchange specifications (22,23). Images from multiple automated microscopes using varied imaging modalities and stains should provide rich training and test datasets.
As our user community is located around the world, all user interaction is via the Internet through standards-compliant web browser pages. All functions are available to authenticated Stanford researchers and their collaborators with authorization to access given experiments governed by experiment to group mappings maintained in the database. Data access is restricted by group until publication, at which time it is made visible to the public. Public data may be searched, the analysis pipeline may be run and both input and output datasets may be freely downloaded along with associated images.
All functions described are available to users who choose to download and install their own instance of the database. For those users who are considering whether to install TMAD for use in their own lab, we have provided a live ‘Demo Mode’ to allow everyone to try the Online Scoring function, as well as online tutorials with screen shots of data management and curator tools.
TISSUE MICROARRAY DESIGN AND CONSTRUCTION
Selecting and laying out the hundreds of donor tissues used per tissue microarray manually requires significant effort. TMAD provides tools to assist the histologist with array block design and construction (Figure 1). Tissues may be selected from existing donor tissues already archived in TMAD, in which case existing Hematoxylin & Eosin (H&E) stained images are available for selecting donors. New tissues are added through batch import of de-identified donor information. TMAD is compliant with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule and discloses for research purposes health information, which has been de-identified by removing all 18 patient identifiers in Policy and Procedures for De-Identification of Protected Health Information and Subsequent Re-Identification, 45 CFR 164.514(a)-(c).
Figure 1.
Each tissue microarray contains hundreds of tissue samples. TMAD provides tools to assist the histologist in selecting tissues by parametric search as well as by providing batch upload of new de-identified tissue metadata.
Per slide data such as antibody concentration, source, pre-treatment, dates and concentration of in situ probes are retained in TMAD and displayed during queries.
TISSUE MICROARRAY EXPERIMENTS
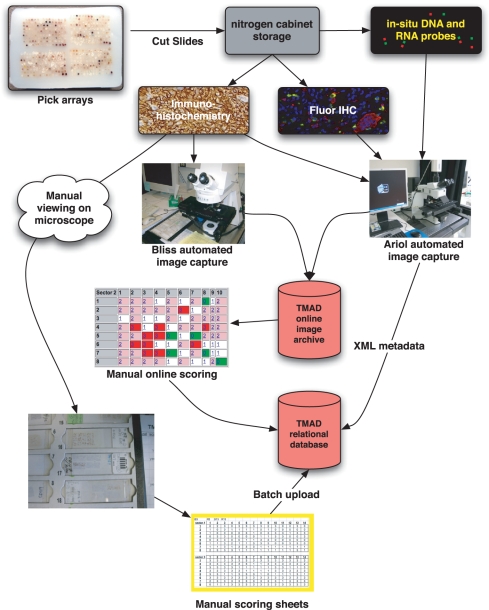
Once the tissue microarrays have been designed, constructed and stained, we use a variety of automated microscopes to permanently archive the resulting images using the typical workflow shown in Figure 2. Immunohistochemistry images of chromogenic and fluorescence secondary probes are captured using one of three microscopes. In each case, the histologist uploads the resulting image and metadata files directly across an encrypted link to a per-user staging area. We use a Bacus Labs Bliss microscope for most chromogenic slides and either a custom in-house automated fluorescence microscope or more recently, an Applied Imaging Ariol bright-field/fluorescence microscope. The histologist sees a browser listing of available slides for import into TMAD.
Figure 2.
Tissue microarray workflow: using TMAD-specific microarray block(s) are selected for an experiment. Slides are cut from the tissue microarray and antibodies are used to visualize protein expression or in situ probes to visualize DNA and RNA targets. The results are imaged on bright-field or fluorescence automated microscopes with results uploaded and archived in TMAD. Pathologists may annotate and score manually or online with results saved for analysis in TMAD.
Many approaches exist for scoring protein, in situ hybridization and FISH experiments. TMAD supports both manual and computer-assisted methods. Direct scoring with a conventional microscope using scoring sheets in spreadsheet format is supported, and the sheets are entered with direct upload via the user's web browser. The spreadsheets are compatible with previous generation tools (24,25), and this allowed us to easily enter hundreds of historical scoring sheets that predated TMAD.
Online scoring is also available and can be used both locally and remotely. While each image is rich in multivariate information, accessing this information requires that the viewer be familiar with histological techniques. The web browser presents a full-size image of the tissue core being scored with buttons to directly enter each score, an annotation area for noting staining localization and an overview sector map. For mixed tissue microarrays, the pathologist can also choose to score all tissues mixed together or separately grouped by tissue type using a pull down with dynamically generated tissue choices.
BROWSING AND SEARCHING TMAD
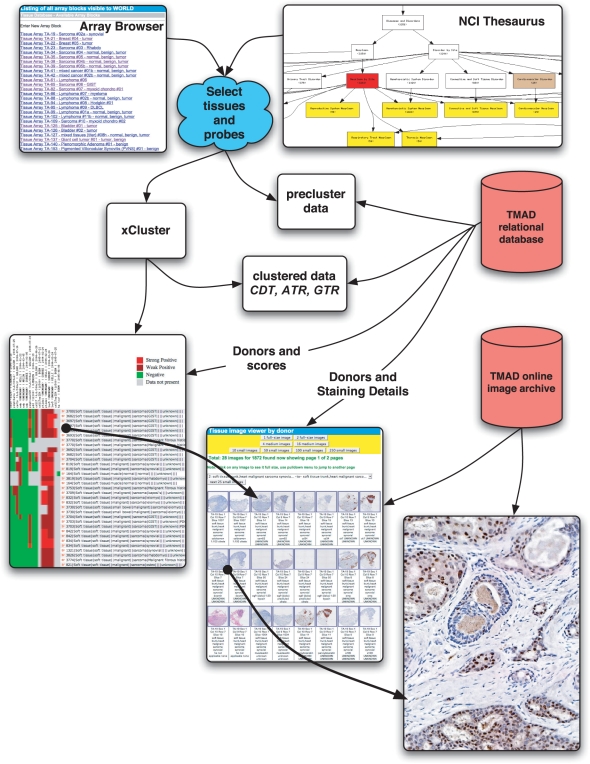
With thousands of stained slides each containing hundreds of individual donors, selection of particular donors requires simple but comprehensive selection tools. As shown in Figure 3, TMAD allows searching for donors at the granularity of an entire microarray or by individual donors through either a controlled vocabulary or traditional informal search terms. It is practical to ask both very general questions such as all breast cancer cases or all ovarian cancer cases, or very specific questions such as all gastrointestinal stromal tumors (GISTs) in stomach but not small bowel.
Figure 3.
TMAD query and analysis tools: tissues and stains are selected by browsing or parametric search, replica spot scores are collapsed and intermediate files are produced for optional download. Data can be clustered, the resulting heatmap visualized and summarized with annotated thumbnail images. Full resolution tissue images are always available by clicking any thumbnail.
Browsing by keyword includes all descriptive tissue sample terms as well as antibody and in situ probe names and common synonyms. As different groups of researchers may use a variety of nomenclatures, we have also designed and implemented tools for mapping tissue metadata into the National Cancer Institute (NCI) Thesaurus (26), which contains over 34 000 concepts arranged as 20 taxonomic trees. The thesaurus provides broad coverage of cancer-related diseases, findings and abnormalities. TMAD provides a graphical browser to the full ontology with clickable links for browsing to more general or specific terms within the NCI trees (27). Our browser shows a live count of the TMAD tissues present by term. Clicking on a term brings up matching stained images.
DATA ANALYSIS PIPELINE
Hierarchical clustering (28) of multiple immunomarkers on tissue microarrays has been demonstrated to be able to group breast cancers into classes with clinical relevance (29) and is also effective for quickly visualizing associations within large datasets. TMAD implements a flexible analysis pipeline including automatic hierarchical clustering. Researchers start by:
Browsing for their tissue, disease or diagnosis of interest as shown in Figure 3.
Selecting donors by tissue, disease or diagnosis on one or several microarrays.
Selecting antibody and/or in situ hybridization probes.
Next, TMAD automatically:
Creates a downloadable annotated pre-cluster data file.
Creates a downloadable tissue microarray data exchange XML file (23) with header, block, slide and core data elements.
Runs XCluster (30) on the annotated data.
Provides downloadable CDT, ATR and GTR post-cluster files.
Creates a graphical clickable heatmap of the clustered data.
Presents the heatmap to the user's web browser.
Colors in the heatmap image represent collapsed scores.
Clicking on a donor row in the heatmap brings up all associated stained images in another browser window for detailed review.
The user may (optionally) download the pre-cluster data for offline analysis. Row and column headings are provided to suit entry into a statistics software package such as R.
The user may (optionally) download the post-cluster files for offline analysis such as preparing high-resolution images for publication using a tool such as Java TreeView (31).
DATABASE AND WEB SERVER SOFTWARE
The Stanford TMAD production server is a SunFire multiprocessor with 4 GB primary memory. All metadata is stored in an Oracle Server Enterprise Edition 9i database. We archive images online in RAID storage arrays with as many bits of image depth as the original source provides. Thumbnail and JPEG versions of the images are pre-computed with ImageMagick for fast viewing over the web. We have tested on current Internet Explorer, Firefox, Safari and Opera web browsers hosted on Windows XP, Mac OS X and Linux platforms and additionally for W3 XHTML 1.0 Transitional compliance.
Web pages are served by Apache 2.0.48 with Perl scripts for querying the database and assembling dynamic output. We have centralized the output style through external CSS. The data analysis pipeline includes Perl scripts and compiled utilities such as XCluster (30). Data on tissues, stains and scores is imported from user spreadsheets through a web browser. On the server side, the imported spreadsheets are processed through the CPAN Spreadsheet module. Source code is available to interested researchers under a no-cost license. Reuse of the source code will be facilitated by familiarity with Oracle, SQL, XHTML, CSS, Perl, the Perl modules CGI, DBI, DBD::Oracle, GD, XML::Simple and Spreadsheet.
TMAD FUTURE DEVELOPMENT
Now that TMAD has met its initial goals of providing a tissue expression database and image archive for Stanford researchers and their collaborators as well as providing online public access to published experiments, we plan on adding additional capabilities. As analyses of greater complexity become the norm, we have found the R statistical computing environment to fit well with tissue microarray analysis tasks and plan on providing additional support for integrating R programs with TMAD. We also hope to extend our design toolset through multiple-choice experiment design pages that produce customized antibody and RNA probe protocols, simultaneously capturing additional experiment metadata.
ACKNOWLEDGEMENTS
Funding for continued development of TMAD is supported by the Howard Hughes Medical Institute (HHMI) and a grant from the National Institutes of Health (5R01 CA77097 to P.O.B.). Funding to pay the Open Access publication charges for this article was provided by HHMI. P.O.B is an investigator of the Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wan W, Fortuna M, Furmanski P. A rapid and efficient method for testing immunohistochemical reactivity of monoclonal antibodies against multiple tissue samples simultaneously. J. Immunol. Methods. 1987;103:121–129. doi: 10.1016/0022-1759(87)90249-3. [DOI] [PubMed] [Google Scholar]
- 2.Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch M, Sauter G, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 3.Lee H, Park Y, Sim J, Park R, Kim W, Kim J. The tissue microarray object model: a data model for storage, analysis, and exchange of tissue microarray experimental data. Arch. Pathol. Lab. Med. 2006;130:1004–1013. doi: 10.5858/2006-130-1004-TTMOMA. [DOI] [PubMed] [Google Scholar]
- 4.Manley S, Mucci N, De Marzo A, Rubin M. Relational database structure to manage high-density tissue microarray data and images for pathology studies focusing on clinical outcome: the prostate specialized program of research excellence model. Am. J. Pathol. 2001;159:837–843. doi: 10.1016/S0002-9440(10)61759-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thallinger G, Baumgartner K, Pirklbauer M, Uray M, Pauritsch E, Mehes G, Buck C, Zatloukal K, Trajanoski Z. TAMEE: data management and analysis for tissue microarrays. BMC Bioinformatics. 2007;8:81. doi: 10.1186/1471-2105-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conway C, O'Shea D, O'Brien S, Lawler D, Dodrill G, O'Grady A, Barrett H, Gulmann C, O'Driscoll L, et al. The development and validation of the Virtual Tissue Matrix, a software application that facilitates the review of tissue microarrays on line. BMC Bioinformatics. 2006;7:256. doi: 10.1186/1471-2105-7-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demichelis F, Sboner A, Barbareschi M, Dell'Anna R. TMABoost: an integrated system for comprehensive management of tissue microarray data. IEEE Trans. Inf. Technol. Biomed. Publ. IEEE Eng. Med. Biol. Soc. 2006;10:19–27. doi: 10.1109/titb.2005.855540. [DOI] [PubMed] [Google Scholar]
- 8.Kim R, Demichelis F, Tang J, Riva A, Shen R, Gibbs D, Mahavishno V, Chinnaiyan A, Rubin M. Internet-based Profiler system as integrative framework to support translational research. BMC Bioinformatics. 2005;6:304. doi: 10.1186/1471-2105-6-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma-Oates A, Quirke P, Westhead D. TmaDB: a repository for tissue microarray data. BMC Bioinformatics. 2005;6:218. doi: 10.1186/1471-2105-6-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vrolijk H, Sloos W, Mesker W, Franken P, Fodde R, Morreau H, Tanke H. Automated acquisition of stained tissue microarrays for high-throughput evaluation of molecular targets. J. Mol. Diagn. 2003;5:160–167. doi: 10.1016/S1525-1578(10)60468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uhlén M, Björling E, Agaton C, Szigyarto C, Amini B, Andersen E, Andersson A, Angelidou P, Asplund A, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Uhlen M, Ponten F. Antibody-based proteomics for human tissue profiling. Mol. Cell. Proteomics. 2005;4:393. doi: 10.1074/mcp.R500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Vyberg M, Torlakovic E, Seidal T, Risberg B, Helin H, Nielsen S. Nordic immunohistochemical quality control. Croat Med. J. 2005;46:368–371. [PubMed] [Google Scholar]
- 14.Gratzinger D, Zhao S, Marinelli R, Kapp A, Tibshirani R, Hammer A, Hamilton-Dutoit S, Natkunam Y. Microvessel density and expression of vascular endothelial growth factor and its receptors in diffuse large B-cell lymphoma subtypes. Am. J. Pathol. 2007;170:1362–1369. doi: 10.2353/ajpath.2007.060901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins J, Kaygusuz G, Wang L, Montgomery K, Mason V, Zhu S, Marinelli R, Presti J, van de Rijn M, et al. Placental S100 (S100P) and GATA3: markers for transitional epithelium and urothelial carcinoma discovered by complementary DNA microarray. Am. J. Surg. Pathol. 2007;31:673–680. doi: 10.1097/01.pas.0000213438.01278.5f. [DOI] [PubMed] [Google Scholar]
- 16.Natkunam Y, Vainer G, Chen J, Zhao S, Marinelli R, Hammer A, Hamilton-Dutoit S, Pikarsky E, Amir G, et al. Expression of the RNA-binding protein VICKZ in normal hematopoietic tissues and neoplasms. Haematologica. 2007;92:176–183. doi: 10.3324/haematol.10724. [DOI] [PubMed] [Google Scholar]
- 17.Natkunam Y, Zhao S, Mason D, Chen J, Taidi B, Jones M, Hammer A, Hamilton Dutoit S, Lossos I, et al. The oncoprotein LMO2 is expressed in normal germinal-center B cells and in human B-cell lymphomas. Blood. 2007;109:1636–1642. doi: 10.1182/blood-2006-08-039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian S, West R, Marinelli R, Nielsen T, Rubin B, Goldblum J, Patel R, Zhu S, Montgomery K, et al. The gene expression profile of extraskeletal myxoid chondrosarcoma. J. Pathol. 2005;206:433–444. doi: 10.1002/path.1792. [DOI] [PubMed] [Google Scholar]
- 19.West R, Rubin B, Miller M, Subramanian S, Kaygusuz G, Montgomery K, Zhu S, Marinelli R, De Luca A, et al. A landscape effect in tenosynovial giant-cell tumor from activation of CSF1 expression by a translocation in a minority of tumor cells. Proc. Natl Acad. Sci. USA. 2006;103:690–695. doi: 10.1073/pnas.0507321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Natkunam Y, Lossos I, Taidi B, Zhao S, Lu X, Ding F, Hammer A, Marafioti T, Byrne G, et al. Expression of the human germinal center-associated lymphoma (HGAL) protein, a new marker of germinal center B-cell derivation. Blood. 2005;105:3979–3986. doi: 10.1182/blood-2004-08-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bray T, Paoli J, Sperberg-McQueen CM, Maler E, Yergeau F, editors. Extensible Markup Language (XML) 1.0. 4th edn. 2006. World Wide Web Consortium [cited 2007 July 16]. Available from: http://www.w3.org/TR/REC-xml/
- 22.Berman J, Datta M, Kajdacsy-Balla A, Melamed J, Orenstein J, Dobbin K, Patel A, Dhir R, Becich M. The tissue microarray data exchange specification: implementation by the Cooperative Prostate Cancer Tissue Resource. BMC Bioinformatics. 2004;5:19. doi: 10.1186/1471-2105-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berman J, Edgerton M, Friedman B. The tissue microarray data exchange specification: a community-based, open source tool for sharing tissue microarray data. BMC Med. Inform. Decis. Mak. 2003;3:5. doi: 10.1186/1472-6947-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C, Montgomery K, Natkunam Y, West R, Nielsen T, Cheang M, Turbin D, Marinelli R, van de Rijn M, et al. TMA-Combiner, a simple software tool to permit analysis of replicate cores on tissue microarrays. Mod. Pathol. 2005;18:1641–1648. doi: 10.1038/modpathol.3800491. [DOI] [PubMed] [Google Scholar]
- 25.Liu C, Prapong W, Natkunam Y, Alizadeh A, Montgomery K, Gilks C, van de Rijn M. Software tools for high-throughput analysis and archiving of immunohistochemistry staining data obtained with tissue microarrays. Am. J. Pathol. 2002;161:1557–1565. doi: 10.1016/S0002-9440(10)64434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Coronado S, Haber MW, Sioutos N, Tuttle MS, Wright LW. NCI Thesaurus: using science-based terminology to integrate cancer research results. Medinfo. 2004;11:33–37. [PubMed] [Google Scholar]
- 27.Shah N, Rubin D, Supekar K, Musen M. Ontology-based Annotation and Query of Tissue Microarray Data. AMIA Annu. Symp. Proc. 2006;2006:709–713. [PMC free article] [PubMed] [Google Scholar]
- 28.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makretsov N, Huntsman D, Nielsen T, Yorida E, Peacock M, Cheang M, Dunn S, Hayes M, van de Rijn M, et al. Hierarchical clustering analysis of tissue microarray immunostaining data identifies prognostically significant groups of breast carcinoma. Clin. Cancer Res. 2004;10:6143–6151. doi: 10.1158/1078-0432.CCR-04-0429. [DOI] [PubMed] [Google Scholar]
- 30.Sherlock G. Analysis of large-scale gene expression data. Curr. Opin. Immunol. 2000;12:201–205. doi: 10.1016/s0952-7915(99)00074-6. [DOI] [PubMed] [Google Scholar]
- 31.Saldanha A. Java Treeview – extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]