Abstract
IMG/M is a data management and analysis system for microbial community genomes (metagenomes) hosted at the Department of Energy's (DOE) Joint Genome Institute (JGI). IMG/M consists of metagenome data integrated with isolate microbial genomes from the Integrated Microbial Genomes (IMG) system. IMG/M provides IMG's comparative data analysis tools extended to handle metagenome data, together with metagenome-specific analysis tools. IMG/M is available at http://img.jgi.doe.gov/m
INTRODUCTION
Studies of the collective genomes (also known as metagenomes) of environmental microbial communities (also known as microbiomes) are expected to lead to advances in environmental cleanup, agriculture, industrial processes, alternative energy production and human health (1). Metagenomes of specific microbiome samples are sequenced by organizations worldwide such as the Department of Energy's (DOE) Joint Genome Institute (JGI), the Venter Institute, Washington University in St. Louis, and Genoscope using different sequencing strategies, technology platforms and annotation procedures. According to the Genomes OnLine Database, about 25 metagenome studies have been published to date, with over 60 other projects ongoing and more in the process of being launched (2). JGI is one of the major contributors of metagenome sequence data, currently sequencing more than 50% of the reported metagenome projects worldwide.
Due to the higher complexity, inherent incompleteness and lower quality of metagenome sequence data, traditional assembly, gene prediction and annotation methods do not perform as well on these datasets as they do on isolate microbial genome sequences (3,4). In spite of these limitations, metagenome data are amenable to a variety of analyses, as illustrated by several recent studies (5–10). Metagenome data analysis is usually set up in the context of reference isolate genomes and considers the questions of phylogenetic composition and functional or metabolic potential of individual microbiomes, as well as differences between microbiome samples. Such analysis relies on efficient management of genome and metagenome data collected from multiple sources, while taking into account the iterative nature of sequence data generation and processing.
IMG/M aims at providing support for comparative metagenome analysis in the integrated context of microbial genome and metagenome data generated with diverse sequencing technology platforms and data processing methods. IMG/M was initially developed as an experimental system (11). Subsequently, IMG/M has been extended in terms of metagenome data content and metagenome specific analytical tools, as discussed below.
DATA CONTENT
IMG/M consists of microbial metagenome data integrated with isolate microbial genomes from the Integrated Microbial Genomes (IMG) system (12). The current version of IMG/M (as of September 2007) contains metagenome datasets generated using shotgun sequencing for 10 projects involving a total of 24 microbiome samples, including an acid mine drainage biofilm (5), three isolated deep sea ‘whale fall’ carcass samples, an agricultural soil sample (6), two biological phosphorus removing sludge samples (7), the metagenome of gutless marine worm symbionts (8), two human distal gut samples (9) and obese and lean mouse gut samples (10). Several other metagenome datasets such as hypersaline microbial mats and termite hindgut metagenomes, are currently analyzed using an internal version of IMG/M in preparation for publication.
The current version of IMG/M also includes 2301 genomes from IMG 2.0 (released on 1 December 2006), consisting of 595 bacterial, 32 archaeal, 13 eukaryotic and 1661 phage genomes.
Similar to IMG, the data model underlying IMG/M allows recording the primary sequence information and its organization in scaffolds and/or contigs, together with computationally predicted protein-coding sequences and some RNA-coding genes. Protein-coding genes are characterized in terms of additional annotations such as motifs, domains, pathways and orthology relationships, which may serve as an indication of their functions. These annotations are based on diverse data sources such as COG (13), Pfam (14) and KEGG (15). Genes are assigned to COGs and Pfams based on reverse position specific BLAST (RPS-BLAST) and NCBI's Conserved Domain Database (16). Homologs are computed as unidirectional hits with an E-value of 10−2 or better, with IMG/M providing support for filtering homolog lists by percent identity, bit score and more stringent E-values.
Isolate organisms are identified via their taxonomic lineage (domain, phylum, class, order, family, genus, species and strain), while individual microbiome samples are treated as ‘meta’ organisms. The sequences of a microbiome sample together with their associated genes and annotations are grouped into ‘bins’ when binning has been performed to assign these sequences to organism types (phylotypes). Both isolate organisms and microbiome samples are characterized by a variety of metadata attributes. Some metadata such as phenotype, habitat, disease, relevance, temperature and pH, are included from GOLD (2), with additional metadata collected directly from scientists or publications.
DATA ANALYSIS
We briefly review below the IMG/M data analysis tools with emphasis on the support for new metagenome analysis tools developed since IMG/M's initial public release in 2006 (11).
Data exploration and visualization tools
Data exploration tools in IMG/M help selecting and examining genomes/metagenomes, genes and functions of interest. Similar to IMG, genes and functions can be selected using keyword searches or functional classification (e.g. COG, Pfam) browsers. Lists of genes and functional annotations of interest can be maintained and further explored using various ‘Analysis Carts’.
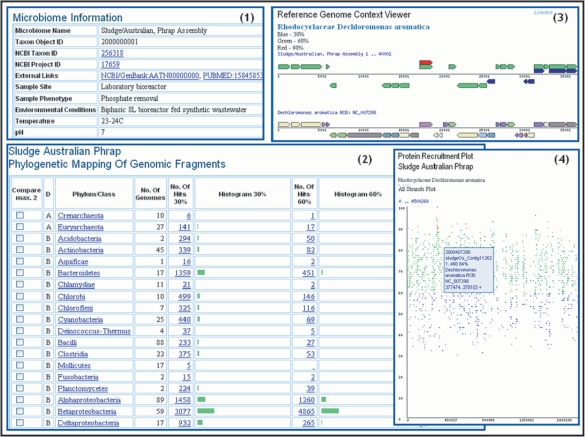
Metagenomes and isolate genomes can be selected using a keyword based ‘Genome Search’ tool or a ‘Genome Browser’. Microbiomes can be further examined using the ‘Microbiome Details’, where a user can find relevant metadata such as sample site, as shown in pane 1 of Figure 1, along with various summaries of interest such as the total number of scaffolds and genes or the number of genes associated with functional characterizations (e.g. COG, Pfam). The ‘Phylogenetic Distribution of Genes’, shown in pane 2 of Figure 1, provides an estimate of the phylogenetic composition of a microbiome sample based on the distribution of the best BLAST hits of the protein-coding genes in the sample. The ‘Phylogenetic Distribution of Genes’ consists of a histogram, with counts of protein-coding genes in the sample that have best BLASTp hits to proteins of isolate genomes in each phylum or class with more than 90% identity (right column), 60–90% identity (middle column) and 30–60% identity (left column). The higher the number of hits and percent identity cutoff, the more likely it is that the sample contains close relatives of the sequenced isolate genomes from this phylum/class. Gene counts in the histogram are linked to the corresponding lists of genes, which can then be selected and added to ‘Gene Cart’ or analyzed through their ‘Gene Pages’. For each phylum/class, the phylogenetic distribution of genes can be projected onto the families in that phylum/class; for each family the distribution of genes can be further projected onto the species in that family. Finally, the genes in the sample can be viewed in the context of an individual reference isolate genome, using either the ‘Reference Genome Context Viewer’ as shown in pane 3 of Figure 1, or the ‘Protein Recruitment Plot’, as shown in pane 4 of Figure 1. The ‘Reference Genome Context Viewer’ displays the metagenome genes aligned with their homologous genes of the reference isolate genome. The metagenome genes are color coded to indicate BLAST percent identity (blue for 30%, green for 60% and red for 90%), while the genes of the reference genome are color coded to indicate their COG functional role, and are displayed as they are located along the chromosome. The ‘Protein Recruitment Plot’ displays the BLASTp hits of the metagenome genes against the genes of the reference genome, with the coordinates of the scaffold reference genome and the BLAST percent identities shown on the X and Y axis, respectively.
Figure 1.
Metagenome Data Exploration and Visualization Tools. Individual microbiome samples such as the ‘Sludge/Australian’ sample, can be examined using the ‘Microbiome Details’ page, which includes relevant microbiome information (1). The ‘Phylogenetic Distribution of Genes’ tool (2) displays the distribution of best BLAST hits of protein-coding genes in the microbiome as a histogram, with counts of genes that have best BLASTp hits to proteins of isolate genomes in each phylum or class with more than 90% identity, 60–90% identity and 30–60% identity. The distribution of genes for each phylum/class can be projected onto the families in that phylum/class such as Betaproteobacteria, and then further projected onto the species in that family such as Rhodocyclaceae. The genes in the sample can be viewed in the context of an individual reference isolate genome such as Dechloromonas aromatica, using the ‘Reference Genome Context Viewer’ (3), or using a ‘Protein Recruitment Plot’ (4). For each gene on ‘Reference Genome Context Viewer’ and ‘Protein Recruitment Plot’, locus tag and scaffold coordinates are provided locally (by placing the cursor over the gene), while additional information is available in the ‘Gene Details’ page, which is linked to each gene.
Similar to genes of isolate genomes, metagenome genes can be examined using ‘Gene Details’ pages, which include information on locus, biochemical properties of the product, KEGG pathways, as well as evidence for the functional prediction: gene neighborhood, COG and Pfam and precomputed lists of homologs, orthologs and paralogs (for isolate organisms), or intra-metagenome homologs as well as homologs to other genomes and metagenomes (for microbiomes).
For metagenomes that include contigs and scaffolds generated by assembly of individual reads and potentially comprised of sequences from multiple strains, a ‘SNP BLAST’ tool allows to examine the heterogeneity between the reads contributing to the composite population contigs and scaffolds. This tool allows users to run BLASTn of the query nucleotide sequence of a specific gene or scaffold in the metagenome against a database of sequencing reads. The BLAST output, which shows whether there are any SNPs among the reads corresponding to the query sequence, can be examined using the raw BLAST output or using the ‘SNP VISTA’ viewer (17).
Comparative analysis tools
Comparative analysis of genomes and metagenomes is provided in IMG/M through a number of tools that allow to examine their gene content and functional capabilities. The differences in gene content of genomes and metagenomes can be examined with a profile-based selection tool (‘Phylogenetic Profiler’) and further explored through gene neighborhood analysis and multiple sequence alignment tools, which are similar to their IMG counterparts (12). Functional capabilities of a microbial community can be examined using several occurrence and abundance profile-based tools. We discuss below in more detail the abundance profile tools that are specific to metagenome data comparative analysis.
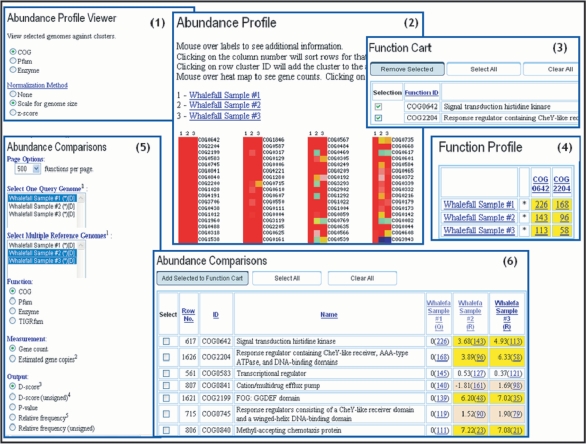
Several ‘Abundance Profile’ tools can be used for comparing the functional capabilities of metagenomes and genomes. The ‘Abundance Profile Viewer’ provides an overview of the relative abundance of protein families (COGs and Pfams) and functional families (enzymes) across selected metagenomes and isolate genomes, as illustrated by the example in pane 1 of Figure 2 which shows the abundance profiles of COGs across three whale-fall microbiomes. Abundance of protein/functional families is displayed as a heat map over all families of a specific type (COGs, Pfams, or enzymes), as shown in pane 2 of Figure 2, with red corresponding to the most abundant families. Each column on the map corresponds to a genome or metagenome, while each row corresponds to a family. Clicking on the cell will retrieve the list of genes assigned to this particular family in this genome or metagenome, while clicking on the identifier of the family displayed on right side of the column (e.g. COG0642) will add the corresponding family to the ‘Function Cart’, as shown in pane 3 of Figure 2. For protein families in the ‘Function Cart’ a selective ‘Function Profile’ can be computed, as shown in pane 4 of Figure 2.
Figure 2.
Abundance Profile Tools. The ‘Abundance Profile Viewer’ (1) provides an overview of the relative abundance of protein families (COGs and Pfams) and functional families (enzymes) across selected metagenomes, normalized for genome size or using z-score. Abundance of protein/functional families is displayed as a heat map (2), with each cell hyperlinked to the list of genes assigned to a particular family. A protein family can be saved in the ‘Function Cart’ by clicking its identifier such as COG0642 (3). For protein families in the ‘Function Cart’ a selective ‘Function Profile’ can be also computed (4). The ‘Abundance Comparison’ tool (5) takes into account the stochastic nature of metagenome datasets and tests whether the differences in abundance can be ascribed to chance variation or not. In addition to the gene count based abundance, the results provided by this tool include an assessment of statistical significance in terms of D-score (6) or P-value.
The ‘Abundance Profile Viewer’ and ‘Function Profile’ tools provide a rough estimate of the functional capabilities of metagenomes. When metagenomes are compared to each other or to isolate genomes, statistical tests are needed for estimating the statistical significance of the observed differences. The ‘Abundance Comparison’ tool, illustrated in pane 5 of Figure 2, takes into account the stochastic nature of metagenome datasets and tests whether the differences in abundance can be ascribed to chance variation or not. The results provided by this tool include an assessment of statistical significance in terms of a d-score (that translates into a P-value) in addition to the gene count based abundance, as shown in pane 6 of Figure 2. The d-score is a standard normal statistics derived under a binomial assumption where the corresponding P-value provides support at different levels of significance (e.g. 0.05, 0.01).
FUTURE PLANS
The current version of IMG/M contains data on 2301 isolate genomes, 21 metagenome samples from 9 studies and 3 simulated datasets from a metagenome data processing benchmarking project (4). New metagenome datasets are continuously included into IMG/M from metagenome studies conducted at JGI and other institutes such as the Washington University in St. Louis, while new reference genomes are included from IMG.
New visualization tools are currently developed in order to improve the efficiency of analyzing large and complex metagenome datasets, including datasets generated with new technology platforms such as the Genome Sequencer 20™ System from 454 Life Sciences. The abundance profile tools will be extended to allow comparison of genomes and metagenomes based on higher-level functional categories such as COG functional categories and KEGG pathways. As the number of analytical tools increases, the organization and documentation of the IMG user interface will be revised in order to improve its usability.
We also plan to extend IMG/M's capability to capture more detailed metadata attributes characterizing microbiome samples. Such attributes are often specific to a habitat (e.g. biomedical, ecological). Samples are associated with properties used for metagenome analysis such as sample structural and morphological characteristics (e.g. sample site, time of collection) and donor or host data (e.g. demographic and clinical record, including diagnosis, disease, stage of disease and treatment information for human donors). Samples may also be involved in clinical studies and therefore can be grouped into several time/treatment study groups. In addition to extending the data model for supporting sample metadata, we plan to improve the coherence and completeness of these annotations via manual curation. We collaborate with the Genome Standards Consortium (18) in order to ensure high coverage and consistency of microbiome sample metadata.
The current version of IMG/M does not provide support for data curation. We plan to incorporate into IMG/M the annotation capabilities that are available in IMG for isolate genomes, adapted to handle metagenome data.
ACKNOWLEDGEMENTS
We thank Chris Oehmen of the Computational Biology and Bioinformatics group at the Pacific Northwest National Laboratory for his help in carrying out the large-scale gene similarity computations for IMG/M. The work of JGI's sequencing, assembly and annotation teams is an essential prerequisite for IMG/M. Eddy Rubin and James Bristow provided, support, advice and encouragement throughout this project. The work presented in this article was performed under the auspices of the US Department of Energy's; Office of Science, Biological and Environmental Research Program and by the University of California, Lawrence Livermore National Laboratory under contract No. W-7405-Eng-48, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231 and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396. Funding to pay the Open Access publication charges for this article was provided by the Department of Energy Joint Genome Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.National Research Council Committee on Metagenomics. The New Science of Metagenomics: Revealing the Secrets of our Microbial Planet. Washington, DC: National Academies Press; 2007. [PubMed] [Google Scholar]
- 2.Liolios K, Tavernarakis N, Hugenholtz P, Kyrpides N. The genomes online database (GOLD) v.2: a monitor of genomeprojects worldwide. Nucleic Acids Res. 2006;34:D332–D334. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen K, Pachter L. Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput. Biol. 2005;1:106–112. doi: 10.1371/journal.pcbi.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mavromatis K, Ivanova N, Barry K, Shapiro H, Goltsman E, McHardy AC, Rigoutsos I, Salamov A, Korzeniewski F, et al. On the fidelity of processing metagenomic sequences using simulated dataset. Nat. Meth. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- 5.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin E, Rokhsar DS, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 6.Tringe S, von Mering C, Kobayashi A, Salamov A, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 7.Martin HG, Ivanova NN, Kunin V, Warnecke F, Barry KW, McHardy AC, Yeates C, He S, Salamov AA, et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 2006;24:1263–1269. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- 8.Woyke T, Teeling H, Ivanova NN, Huntemann M, Richter M, Gloeckner FO, Biffelli D, Anderson I, Barry KW, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- 9.Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 11.Markowitz VM, Ivanova N, Korzeniewski F, Palaniappan K, Szeto E, Lykidis A, Anderson I, Mavrommatis K, Kunin V, et al. An experimental metagenome data management and analysis system. Bioinformatics. 2006;22:e359–e367. doi: 10.1093/bioinformatics/btl217. [DOI] [PubMed] [Google Scholar]
- 12.Markowitz VM, Szeto E, Palaniappan K, Chen IA, Grechkin Y, Chu K, Dubchak I, Anderson I, Lykidis A, et al. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkm846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatusov RL, Koonin EV, Lipman DJA. Genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 14.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah N, Teplitsky MV, Minovitsky S, Pennacchio LA, Hugenholtz P, Hamann B, Dubchak IL. SNP-VISTA: an interactive SNP visualization tool. BMC Bioinformatics. 2005;8:292. doi: 10.1186/1471-2105-6-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Field D, Garrity G, Gray T, Selengut J, Sterk P, Thomson N, Tatusov T, Cochrane G, Glockner FO, et al. eGenomics: cataloguing our complete genome collection III. Comp. Funct. Genomics. 2007;10:100–104. doi: 10.1002/cfg.494. [DOI] [PMC free article] [PubMed] [Google Scholar]