Abstract
Accurate and comprehensive information about the nucleotide sequence specificity of trans-acting factors (TFs) is essential for computational and experimental analyses of gene regulatory networks. We present the Yeast Transfactome Database, a repository of sequence specificity models and condition-specific regulatory activities for a large number of DNA- and RNA-binding proteins in Saccharomyces cerevisiae. The sequence specificities in TransfactomeDB, represented as position-specific affinity matrices (PSAMs), are directly estimated from genomewide measurements of TF-binding using our previously published MatrixREDUCE algorithm, which is based on a biophysical model. For each mRNA expression profile in the NCBI Gene Expression Omnibus, we used sequence-based regression analysis to estimate the post-translational regulatory activity of each TF for which a PSAM is available. The trans-factor activity profiles across multiple experiments available in TransfactomeDB allow the user to explore potential regulatory roles of hundreds of TFs in any of thousands of microarray experiments. Our resource is freely available at http://bussemakerlab.org/TransfactomeDB/
INTRODUCTION
Gene- and condition-specific regulation of transcription rate is mediated by interactions between trans-acting regulatory factors and DNA. Through these protein interfaces to the genome, the cell can tightly control gene expression in response to environmental or developmental signals. If we can predict the affinity with which a nucleotide sequence is bound by a particular regulatory protein, we can make predictions about the extent to which the corresponding gene is subject to regulation by that factor. This knowledge can suggest future experiments and allow for computational analysis of gene expression. The community has therefore long made efforts to discover, collect, organize and present sequence specificity information for DNA-binding proteins (DBPs) (1–4).
The two largest online databases of sequence specificity information are TRANSFAC (5) and JASPAR (6). These databases compile sequences that are known or believed to be bound with high affinity by particular DBPs, derived either by in vitro selection of tightly bound oligonucleotides (SELEX) or by experimental determination of actual transcription factor binding sites. Both databases align the collected sequences and summarize the sequence specificity of DBPs as position weight matrices (PWMs), which summarize how many sequences have a given nucleotide at a given position in the transcription factor binding site. A PWM can be used to define a position-specific scoring matrix (PSSM), whose entries can be related to binding free energies, but only by making by rather strong assumptions about how the rate of evolutionary selection of individual binding sites depends on their relative affinity (1,2).
The advent of DNA microarrays has greatly aided in discovering the nucleotide sequence specificities of trans-factors. Microarrays have been used for measuring the in vivo association of DBPs with upstream promoter regions (7,8), the in vitro association of DBPs with long (9,10) or short (11,12) segments of DNA and the association of RNA-binding proteins (RBPs) with mRNA molecules (13,14). Typically, motif-finding methods based on information theory [see (3) for early examples] are applied to the most strongly bound sequences from these experiments, providing PSSM representations of the binding specificities of the assayed DBPs or RBPs. Recently, MacIsaac et al. (15) created a large collection of PSSMs for DBPs in Saccharomyces cerevisiae from ChIP-chip data using this class of methods.
Unlike traditional low-throughput methods, however, genomewide binding data provides thousands of examples of sequences rather than only a handful, and a numerical value proportional to overall trans-acting factor (TF) occupancy is available for each sequence rather than only the binary distinction between bound and unbound. The information theory-based algorithms do not take full advantage of this quantitative information and therefore may produce sequence specificities that are less accurate than is possible. In particular, binding energies are only inferred up to an unknown scaling factor. Therefore, the resulting PSSMs can only be used to approximately rank candidate TF-binding sites by affinity, not to obtain a quantitative estimate of their relative affinity.
To address this issue, we recently developed the MatrixREDUCE algorithm, which employs a statistical-mechanical model of protein–nucleic acid binding to infer sequence specificities from mRNA expression data (16) or genomewide occupancy data (17). MatrixREDUCE directly integrates microarray intensities with nucleotide sequence data to infer the free energies of sequence-specific protein–nucleic acid interactions. The algorithm represents this information as a position-specific affinity matrix (PSAM; see (17) for a detailed derivation of PSAMs and the MatrixREDUCE model). Briefly, a PSAM is populated with relative affinities for each nucleotide at each position in the binding site that are directly related to the free energy of binding between the protein and the nucleic acid. MatrixREDUCE avoids the problematic assumption of affinity-proportional sequence representation. As a consequence, it does not require a background sequence model. [For a full discussion of these issues, see Bussemaker et al. (4).]
To create the Yeast Transfactome Database, we applied an updated version of MatrixREDUCE to hundreds of available in vivo and in vitro genomewide occupancy datasets for both DNA- and RNA-binding proteins for S. cerevisiae. We produced a PSAM for each individual microarray experiment, so there are often multiple examples of PSAMs for the same DBP or RBP, allowing for internal validation in many cases. Using these PSAMs, we applied the sequence-based regression approach of Foat et al. (16) to infer condition-specific, post-translational regulatory activities for each TF for which a PSAM is available across all yeast mRNA expression profiles available in NCBI Gene Expression Omnibus (GEO) (18). The PSAMs can also be used to predict the relative TF-binding affinity for any arbitrary nucleotide sequence. All this information can be browsed and queried via a web interface. To our knowledge, the Yeast Transfactome Database is the most comprehensive source of sequence specificities for TFs for S. cerevisiae. In addition, it has the advantages of (i) having a uniform, biophysically motivated representation of sequence specificities in the form of PSAMs; (ii) providing condition-specific regulatory information for each PSAM; and (iii) predicting single-nucleotide TF-binding affinity profiles for arbitrary DNA and RNA sequences.
DATABASE GENERATION AND CONTENTS
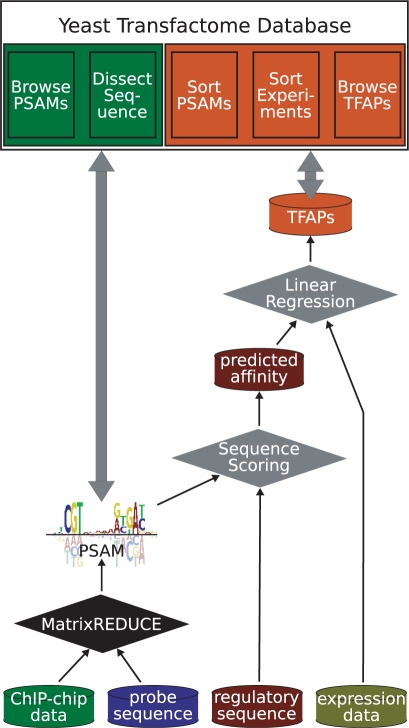
The contents of the Yeast Transfactome Database are original and derived by integrating publicly available microarray and sequence data via computational modelling. The process results in two primary kinds of information: (i) the sequence specificities for trans-factors that have been profiled in genomewide binding assays, and (ii) inferred post-translational regulatory activities for those same TFs in each experimental condition represented by a microarray sample in GEO. Figure 1 illustrates the flow of the low-level primary data through to the derived data types and to the web interfaces through which they can be interrogated. First, genomewide occupancy data and microarray probe sequences were gathered from publication supplements and used as input to the MatrixREDUCE algorithm (16,17). MatrixREDUCE produced a PSAM to represent the sequence specificity of the TF assayed in each experiment. Next, regulatory sequence (upstream promoter regions for DBPs, full-length mRNAs for RBPs) for each gene in the genome was scored for its predicted affinity for the TF represented by the PSAM. Each of thousands of individual microarray experiments from GEO was then regressed on the genomewide profile of gene-specific binding affinity for each PSAM, and the regression coefficient interpreted as a (change in) TF activity in that particular experiment. We refer to the profile of inferred TF activity across all experiments as a trans-factor activity profile (TFAP). The TFAPs represent the majority of the novel results in our database, likely containing hundreds of examples of previously unknown regulatory effects for the assayed TFs.
Figure 1.
The flow of data. Publicly available microarray data and genomic sequence was integrated by MatrixREDUCE and other computational procedures to infer TF sequence specificities (PSAMs) and post-translational regulatory activities (TFAPs). These two data types can be displayed and interrogated using five different ‘tabs’ in the Yeast Transfactome Database interface.
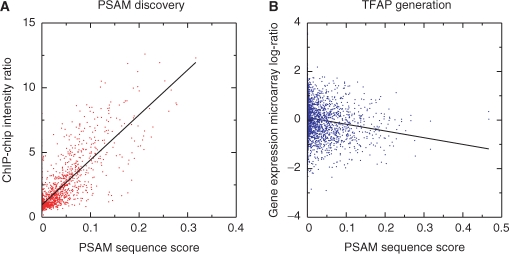
The manner in which PSAMs and TFAPs are generated have similarities and deserve further description to enable a full understanding of the database contents. Both PSAMs and TFAPs result from a model fit that explains microarray intensities in terms of promoter affinities predicted from sequence. For the purposes of this database, however, PSAMs were inferred from genomewide TF binding data through a non-linear fit of the MatrixREDUCE model (Figure 2A). TFAPs were generated by using the discovered PSAMs to predict the total affinity of each regulatory sequence, and then performing linear regression of mRNA expression data on these affinities. The TF activities in each TFAP are represented as regression coefficients scaled by their standard deviations (t-values). Figure 2A shows the fit of ChIP-chip data to the predicted total affinities for an optimized PSAM as would be performed by MatrixREDUCE. Figure 2B shows the fit of data from a particular gene expression microarray experiment to the predicted affinities for the same PSAM, as would be necessary to calculate an entry in its TFAP.
Figure 2.
Regressing microarray data on genomic sequence. (A) Inferring a PSAM from a ChIP-chip experiment. Shown is the result for the transcription factor Abf1p. The parameters of the PSAM are chosen so as to maximize the correlation between chromatin enrichment ratios and the total affinities of the promoter region across all genes. (B) Regression of the change in mRNA expression value on total promoter affinity predicted using a previously computed PSAM can be used to infer changes in the regulatory activity (the slope of the regression line) of the TF whose sequence specificity is represented by the PSAM. In this example, it is shown that between rich media and media containing copper sulphate (GEO accession number GSM17192) mRNA expression levels are downregulated in proportion to the affinity of the promoter region for Abf1p.
At present, the Yeast Transfactome Database contains 399 and 20 automatically-generated PSAMs for 194 DBPs and 5 RBPs, respectively. It also contains TFAPs for each PSAM across more than 4000 microarray experiments. For comparison, MacIsaac et al. (15) provide 124 curated matrices with one matrix per factor, TRANSFAC (Release 10.3) (5) provides 40 matrices for 31 DBPs, and JASPAR (6) has no matrices for S. cerevisiae. Of the 399 DBP PSAMs in the Yeast Transfactome Database, 100 PSAMs corresponding to 52 DBPs are most similar to a PSSM from MacIsaac et al. (15) with the same factor identity. An additional 28 PSAMs corresponding to 11 DBPs are most similar to a PSSM from MacIsaac et al. (15) corresponding to a protein with which the analysed factor is believed to have a physical or genetic association. Physical associations are significant in that an assayed factor may be bound to its target sequences indirectly via protein–protein interactions with a DBP. Genetic associations are significant as factors may have genetic interactions if they have the same sequence specificity (e.g. Msn2p and Msn4p). Fifty-nine PSAMs for 18 proteins are most similar to the TRANSFAC (5) matrix of the same protein identity, and six PSAMs for four proteins are most similar to a matrix for a protein with which the assayed factor has a physical or genetic interaction. In the web interface, it is possible to view only those PSAMs that are consistent with one or both of the other sources of sequence specificities.
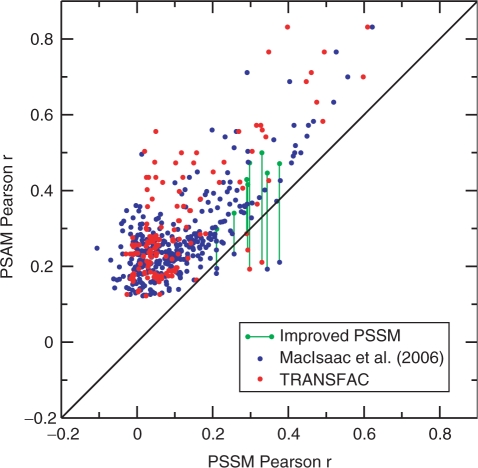
Since MatrixREDUCE is based on a biophysical model of DBP–DNA interactions, and since PSAMs are derived by a direct fit of the model to a particular dataset, a PSAM should always do at least as well at explaining genomewide occupancy measurements as a ‘pseudo-PSAM’ (see Methods section and Supplementary Data) derived from the PWM for the same DBP. We tested this assertion by converting all PWMs from TRANSFAC (5) and MacIsaac et al. (15) to pseudo-PSAMs and comparing to the PSAMs inferred by MatrixREDUCE (Figure 3). In 471 of 480 comparisons, the latter better explained the data, as expected. In the nine cases where the pseudo-PSAM performed better than the true PSAM, an even better fit to the data was achieved by allowing MatrixREDUCE to improve the pseudo-PSAM through the PSAM fitting procedure. Those few cases where the the pseudo-PSAMs performed better were likely due to MatrixREDUCE settling on a suboptimal local minimum.
Figure 3.
Comparison with weight matrices from MacIsaac et al. (15) and TRANSFAC. Each weight matrix from MacIsaac et al. (15) or TRANSFAC (5) was converted into a pseudo-PSAM (see Methods). The correlation between the total affinity of each promoter region predicted by the pseudo-PSAM and the fold-enrichment in the ChIP-chip experiment was then computed. These Pearson r values were then compared with the Pearson r values achieved by PSAMs optimized for the same ChIP-chip data by MatrixREDUCE. In all but nine instances, the correlations were better for PSAMs fit by MatrixREDUCE than for pseudo-PSAMs. In those cases where the pseudo-PSAM had a higher correlation, MatrixREDUCE could still improve the fit of the pseudo-PSAM (green lines).
WEB INTERFACE
The web interface to the Yeast Transfactome Database is available at http://bussemakerlab.org/TransfactomeDB/. The user may choose between examining DNA- or RNA-binding proteins. At the time of writing, genomewide binding data was only publicly available for the five members of the Puf family of RBPs in yeast, so the DBP section of the database is more substantial, covering hundreds of yeast transcription factors.
The main interface consists of five different ‘tabs’ that allow the user to view different kinds of data. Displayed in the ‘Browse PSAMs’ tab are all PSAMs (one for each available genomewide occupancy experiment) along with the name of the TF assayed and the citation for the source of the data. A button at the top of the page allows the user to toggle between two different views of the PSAM list. The first view sorts all PSAMs by their goodness of fit (r2) to the original occupancy data. The t-value provides a guide for the highest quality PSAMs: the higher the t-value, the better the PSAM explained the original data. The other view sorts all PSAMs alphabetically by the common gene name of the TF. Clicking on an affinity logo allows the user to view the actual relative affinities that constitute the PSAM and other details about the PSAM. The user may click on any gene name to look up the TF in the Saccharomyces Genome Database (19).
In the ‘Sort Experiments’ tab, the user is presented with the list of all PSAMs similar to the display from the ‘Browse PSAMs’ tab. Clicking on a PSAM logo presents a list of all experiments, sorted by the absolute t-value of the correlation between gene expression values and predicted regulatory sequence affinities. The user may investigate the experimental design of any of the samples with which she is unfamiliar by clicking on the ‘GEO’ link next to the sample title, which goes to the NCBI GEO website (18).
In the ‘Sort PSAMs’ tab, the user is presented with a list of all parsable (see Methods) gene expression ‘samples’ available in GEO for S. cerevisiae. The samples are listed in order of their GEO sample IDs and are labelled with their GEO sample titles. The user can view the sample information in GEO by clicking on the ‘GEO’ link across from the sample title. Clicking on a link for a sample presents the user with a list of PSAMs sorted by the goodness of fit (absolute t-value) of their predicted promoter occupancy (17) to the expression values in the experiment of interest. Also shown are P-values corresponding to the t-values, and E-values, which are P-values corrected for the number of PSAMs that have been sorted.
The ‘Visualize TFAPs’ tab is useful for visually inspecting patterns of condition-specific regulation for multiple TFs across multiple microarray experiment conditions. It presents this TFAP information in colour matrix displays that were originally developed for visualizing microarray data (20) and later adapted to visualizing TFAPs (16). This interface allows the user to dynamically generate a blue and yellow colour matrix displaying the TFAPs for user-selected PSAMs and experiments. Each coloured box in this display represents the t-value for the fit of the predicted promoter occupancies for the TF (given its PSAM) to the measured expression values for the respective experiment.
The ‘Dissect Sequence’ tab allows the user to view high affinity binding sites for user-selected PSAMs in the preloaded regulatory sequences or in a user-supplied nucleotide sequence. After indicating the desired sequence and PSAMs, the user is presented with a graphical display of the sequence. Any sequence window that has an affinity that would cause the site to be bound at least 5% as strongly as the best binding site in the genome is marked by a coloured box. The stronger the binding site, the darker the box.
CAVEATS
As with any tool in biology, the information in TransfactomeDB should not be interpreted in isolation. However, when the information in the database is combined with knowledge gleaned from the original microarray publications and other literature sources, the user can derive specific and meaningful conclusions. Moreover, the database can be used to generate experimentally testable hypotheses about transcriptional regulation (DBPs) and post-transcriptional regulation (RBPs). Of course, it inherits any limitations of the original data from which it is derived. First, not all microarray experiments in GEO contain meaningful data or necessarily measure only the phenomenon that the experiment was designed to measure. Second, a PSAM derived from genomewide occupancy data is not necessarily the PSAM for the named, assayed factor or even a real PSAM. We report the best fit PSAM for each ChIP-chip or similar experiment. Therefore, if the original occupancy data is due to the specificity of another factor, then the reported PSAM will not accurately reflect the binding specificities for the factor of interest. The t-value for the goodness of fit for the PSAM to the occupancy data can provide a measure of whether the PSAM is derived from pure noise. To mitigate uncertainty in DBP identity, the database allows the user to optionally list only those PSAMs that have a similar predicted occupancies as PSSMs from MacIsaac et al. (15) or PWMs from TRANSFAC (5) for the same DBP or related DBPs. However, there is no computational solution to perfectly prevent reporting PSAMs for factors that are physically interacting with to the immunoprecipitated factor in ChIP-chip experiments. Only data generated from in vitro genomewide occupancy experiments (9–12) can assure that the microarray signal and thus the PSAM is due to the assayed factor. Finally, a high scoring PSAM match for the ‘Dissect Sequence’ tab can only imply potential regulation. This feature can be powerful if combined with previous knowledge and experimental validation. However, the user should expect frequent false positives.
FINAL THOUGHTS
We expect that there are hundreds of cases of biologically interesting differential regulation of TF activity in specific conditions waiting to be uncovered using the Yeast Transfactome Database. The web interface allows the user to dynamically interact with published microarray data and easily discern otherwise hidden regulatory patterns. In addition, all sequence specificity models (PSAMs) were inferred from genomewide occupancy data and are original to the database. TransfactomeDB remains under development, and can in principle be extended to any organism for which comprehensive TF binding and mRNA expression data is available. However, we believe that this first version will already be useful to the community.
METHODS
MatrixREDUCE implementation and parameters
We used the MatrixREDUCE algorithm as described in (16,17) to find the best fit PSAM that could be produced from each microarray experiment. The parameters for all runs of MatrixREDUCE were as follows: the length of each of the two dyads of the seed motifs was three, the length of the added flanks on each side of the dyad was three, the minimum gap was zero, the maximum gap was 15, the minimum allowed relative affinity for any nucleotide was 10− 3. To factor out any nucleotide composition biases in the microarray data (21), a model with regression coefficients for the count on each individual nucleotide was fit to each dataset before fitting a PSAM.
Empirical P-value estimation
The quantity that is maximized by MatrixREDUCE is the absolute value of the Pearson correlation r between the sequence-predicted and actual measured microarray intensities. Because of the non-linear dependence of the PSAM parameters (see Supplementary Data), the null distribution of |r| is not known analytically, and the r2 values obtained for random data are typically much larger than for standard linear regression. Without proper measures, this would give rise to incorrect estimation of the statistical significance of r2. However, we have found that the null distribution can be determined empirically by executing repeated trials of MatrixREDUCE on randomly generated nucleotide sequence and microarray data. We performed approximately 1000 trials for several combinations of parameters defining the randomized data. For each setting of these parameters, the empirical distribution of the Pearson |r| was well approximated by the normal distribution. We found that the mean of the distribution was dependent only on N, the number of sequence-measurement pairs and Lw, the number of optimized nucleotide positions in the PSAM. In addition, we observed that the standard deviation of the empirical distribution of |r| depends solely on the size of the dataset N and is inversely proportional to its square root. The microarray data distributions sampled to reach this conclusion were the following: Gaussian (normal) distribution; skewed Gaussian (all values greater than zero doubled); mixture of Gaussians (90% with SD=1, 10% with SD=2); uniform (rectangular) distribution (an extreme case); permuted actual biological data (a realistic case). The distribution of |r| was also determined to be independent of the lengths of the sequences over a wide range, from ∼200 to 2000 bases. In addition, we found that the distribution of |r| does not depend on the overall base composition statistics of the sequence data, based upon trials using both randomized and true biological sequences.
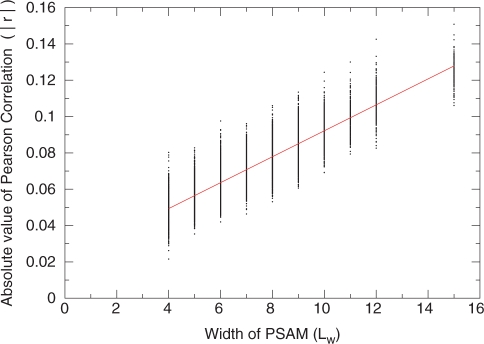
Combining the observations from the above trials and performing linear regression on PSAM width Lw (Figure 4), the estimator of the mean of |r| under the null distribution as a function of Lw and the number of genes N is given by:
| (1) |
while the standard deviation is given by:
| (2) |
Thus, a (pseudo-) t-value corresponding to the Pearson correlation r for a MatrixREDUCE optimized PSAM is:
| (3) |
Since N > 1000 for all of the data analysed here, the corresponding P-value can be well estimated using a standard normal distribution. We used the empirical |r| distribution to calculate t-values and P-values for every PSAM in the database. Only those PSAMs with P-values better than 10− 3 were included.
Figure 4.
Determining the parameters of the empirical P-value calculation for MatrixREDUCE quality of fit. Shown in black are the value of |r|, the absolute value of the Pearson correlation for randomized data at N = 6 505 genes and a range of PSAM widths Lw. The red line shows the result of a linear fit to the data, which gives rise to the results shown in Equation 1.
Genomewide TF binding data
Genomewide occupancy data was gathered from publication supplements. ChIP-chip data for transcription factors was from (7,8,22–25), in vitro protein binding microarray data was from (9) and in vitro DNA immunoprecipitation data was from (10). The affinity selection microarray for the Puf RBPs was from (14). All data was analysed as the ratio of binding-enriched signal versus control signal. All microarray data was purged of extreme outliers before analysis (Grubbs test (26) P-value ≤ 10− 10).
Sequence data
When analysing genomewide occupancy data with MatrixREDUCE, the most accurate results will be obtained when the input sequence set corresponds to the actual bound sequences that give rise to each spot signal on the microarray. In the case of the ChIP-chip, PBM and DIP-chip data, we used the probe sequences themselves as a proxy for the chromatin or DNA fragments that were bound by the DBPs. For the affinity selection microarrays for the RBPs, we used approximated full length mRNA sequences as follows: David et al. (27) measured the mRNA levels for every yeast gene using a genome tiling microarray. Thus, they created a nucleotide-resolution map of the transcriptome expressed under log-growth conditions. While, the data does not contain a measurement for every gene, we used this data to produce approximate full-length mRNA sequences for about half of all yeast genes. We used the fixed 5′ and 3′ UTR lengths for the unknown half of mRNA sequences that gave a 25% per nucleotide false negative rate for the UTRs of the known half.
mRNA expression data
All S. cerevisiae mRNA expression data available in the Gene Expression Omnibus (18) was downloaded and parsed. Datasets were used for further analysis if it was possible to resolve spot IDs to open reading frame (ORF) identifiers and if they contained values for over 4000 ORFs. This resulted in 201 of 228 available data series, which provided 4094 experiments for analysis. All data was analysed in the numerical form that was entered into GEO. These values are often log2-ratios but may be measurements of absolute expression in some instances.
Trans-factor activity profiles
The PSAMs were used to infer regulatory activities associated with their nucleotide specificities. For DBPs, the occupancy was predicted (17) for the 800 bp upstream of every yeast ORF as approximate promoter regions. The predicted occupancies were then correlated with each mRNA expression microarray experiment dataset. The strength and direction of the correlation between predicted DBP binding and mRNA expression was reported as a t-value. The same process was performed for RBPs except that occupancy was predicted over real or approximate full length mRNA sequences rather than approximate promoter regions.
Comparing PSAMs and PSSMs
If we assume that the sequences that give rise to PSSMs or PWMs are represented proportionally to their affinity, we can convert these other matrix formats to ‘pseudo-PSAMs’ (see Supplementary Data). This enables the predictions of true PSAMs and the PSSM- or PWM-derived pseudo-PSAMs to be directly compared. Using these relationships, we converted all of the specificity matrices for S. cerevisiae from TRANSFAC (Release 10.3) (5) and MacIsaac et al. (15) into pseudo-PSAMs. JASPAR (6) has no matrices for S. cerevisiae.
Next we calculated occupancies for each real PSAM and pseudo-PSAM across all intergenic sequences from S. cerevisiae as previously described (17). Pearson correlations were then calculated between the predicted occupancies of the same regions for each pair of PSAMs. We used the resulting r2 values to identify which PSAMs from the Yeast Transfactome best matched matrices from other sources. For each Yeast Transfactome PSAM, we identified the best correlating pseudo-PSAM from both TRANSFAC (5) and MacIsaac et al. (15). We then noted whether the pseudo-PSAM corresponded to the same DBP or an associated factor. Physical or genetic associations between factors were identified using BioGRID (28).
Finally, we used the predicted occupancies of the Yeast Transfactome PSAMs and the pseudo-PSAMs to compare their abilities to explain genomewide occupancy data. For each PSAM, we calculated the Pearson r between the predicted occupancies and the measured intensity ratios values for the genomewide occupancy experiment from which the PSAM was derived. For each pseudo-PSAM, we calculated the Pearson r between the predicted occupancies and the measured intensity ratios for each genomewide occupancy experiment performed for the same DBP.
Website implementation
The Yeast Transfactome Database runs on a Linux, Apache, MySQL, Perl platform. The HTML forms are generated with the help of CGI.pm. Graphics are created using GD.pm. TFAP clustering is accomplished with the help of Algorithm::Cluster.pm (29). The interface with the MySQL database is accomplished via DBI.pm. Affinity logos for PSAMs were generated as previously described (17).
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health (NIH) training grant T32GM008798 (B.C.F.) and grants R01HG003008 and U54CA121852 (to H.J.B.). We thank Gabor Halasz and Lucas Ward for critical readings of the article. Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berg OG, vonHippel PH. Selection of dna binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J. Mol. Biol. 1987;193:723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- 2.Stormo GD, Fields DS. Specificity, free energy and information content in protein-dna interactions. Trends Biochem. Sci. 1998;23:109–113. doi: 10.1016/s0968-0004(98)01187-6. [DOI] [PubMed] [Google Scholar]
- 3.Stormo GD. Dna binding sites: representation and discovery. Bioinform. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 4.Bussemaker HJ, Foat BC, Ward LD. Predictive modeling of genome-wide mrna expression: from modules to molecules. Annu. Rev. Biophys. Biomol. Struct. 2007;36:329–347. doi: 10.1146/annurev.biophys.36.040306.132725. [DOI] [PubMed] [Google Scholar]
- 5.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, et al. Transfac and its module transcompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. Jaspar: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, et al. Genomewide location and function of dna binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 8.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors sbf and mbf. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 9.Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. Rapid analysis of the dna-binding specificities of transcription factors with dna microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Noll DM, Lieb JD, Clarke ND. Dip-chip: rapid and accurate determination of dna-binding specificity. Genome Res. 2005;15:421–427. doi: 10.1101/gr.3256505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren CL, Kratochvil NCS, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips GN, Ansari AZ. Defining the sequence-recognition profile of dna-binding molecules. Proc. Natl Acad. Sci. USA. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, Bulyk ML. Compact, universal dna microarrays to comprehensively determine transcription factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mrna subsets in messenger ribonucleoprotein complexes by using cdna arrays. Proc. Natl Acad. Sci. USA. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mrnas with puf family rna-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for saccharomyces cerevisiae. BMC Bioinform. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foat BC, Houshmandi SS, Olivas WM, Bussemaker HJ. Profiling condition-specific, genomewide regulation of mrna stability in yeast. Proc. Natl Acad. Sci. USA. 2005;102:17675–17680. doi: 10.1073/pnas.0503803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foat BC, Morozov AV, Bussemaker HJ. Statistical mechanical modeling of genomewide transcription factor occupancy data by matrixreduce. Bioinform. 2006;22:e141–e149. doi: 10.1093/bioinformatics/btl223. [DOI] [PubMed] [Google Scholar]
- 18.Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval and analysis. Methods Enzymol. 2006;411:352–369. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Roe T, et al. Sgd: Saccharomyces genome database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of rap1 revealed by genome-wide maps of protein-dna association. Nat. Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 23.Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Gifford DK, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- 24.Zeitlinger J, Simon I, Harbison CT, Hannett NM, Volkert TL, Fink GR, Young RA. Program-specific distribution of a transcription factor dependent on partner transcription factor and mapk signaling. Cell. 2003;113:395–404. doi: 10.1016/s0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 25.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne J, Reynolds DB, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grubbs F. Procedures for detecting outlying observations in samples. Technometrics. 1969;11:1–21. [Google Scholar]
- 27.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc. Natl Acad. Sci. USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. Biogrid: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.deHoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinform. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]