Abstract
Understanding how genetic variation affects the molecular function of gene products is an emergent area of bioinformatic research. Here, we present updates to MutDB (http://www.mutdb.org), a tool aiming to aid bioinformatic studies by integrating publicly available databases of human genetic variation with molecular features and clinical phenotype data. MutDB, first developed in 2002, integrates annotated SNPs in dbSNP and amino acid substitutions in Swiss-Prot with protein structural information, links to scores that predict functional disruption and other useful annotations. Though these functional annotations are mainly focused on nonsynonymous SNPs, some information on other SNP types included in dbSNP is also provided. Additionally, we have developed a new functionality that facilitates KEGG pathway visualization of genes containing SNPs and a SNP query tool for visualizing and exporting sets of SNPs that share selected features based on certain filters.
INTRODUCTION
Understanding how coding single nucleotide polymorphisms (cSNPs) and disease-associated mutations cause molecular alterations and expression changes in gene products is important to many fields of biological and medical research (1,2). We believe that linking disease with basic research data will enable hypothesis generation that can be experimentally tested in the laboratory with functional assays.
Recently, several servers and databases aiming to understand the biochemical effects of nonsynonymous SNPs and disease-associated mutation have been developed. These include SIFT (3), PolyPhen (4), SNPs3D (5), PANTHER (6), PMUT (7), LS-SNP (8), PolyDoms (9) and SNPEffect (10). These methods and their resulting datasets generally apply DNA and protein sequence, protein structure and/or evolutionary features to classify a query amino acid substitution using a training set of putative neutral and causative amino acid substitutions (4,5,8,11–17).
Similarly, MutDB (18,19) is an online resource that serves as a step toward better understanding the potential molecular effects of a mutation. MutDB integrates genetic variation from two public databases, Swiss-Prot (20) and dbSNP (21), and annotates the variants with biochemically relevant information. These two databases are chosen because they are freely available and represent a significant breadth of available amino acid substitutions. However, neither of these databases annotates disease causing amino acid substitutions particularly well. dbSNP contains few links to OMIM (22), and Swiss-Prot does not identify disease causing amino acid substitutions from other amino acid substitutions. Therefore, a researcher studying a specific disease should have some prior knowledge of the proteins and mutations of interest, and MutDB provides some helpful links to useful databases with disease and phenotype annotations such as OMIM and dbGAP (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap), (22).
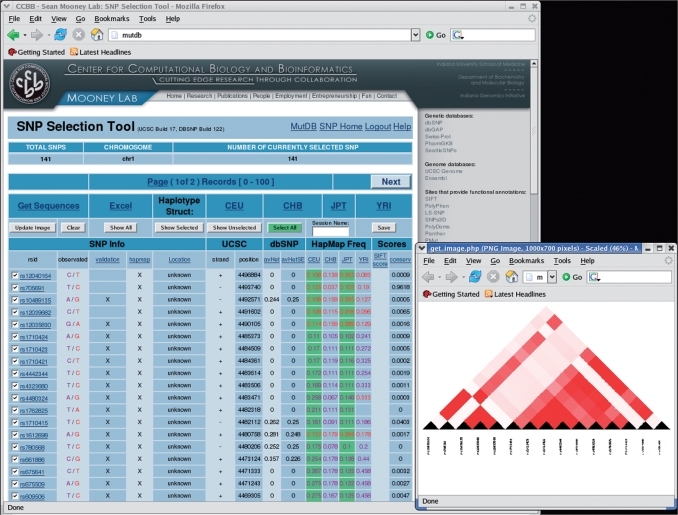
In addition to updating to the latest mutation and SNP datasets, here we present several additions to the MutDB resource. First, we have developed a pathway visualization add-on to MutDB that leads the biologist from mutations in a gene to KEGG (23) biological pathways involving the gene. This enables the researcher to view the systems context of both a mutation and its associated phenotype. Second, we have constructed an AJAX (Asynchronous JavaScript and XML) based SNP query tool that allows users to save searches, view Haploview-like haplotype structure (24), and select subsets of SNPs based on frequencies and SNP scores. Together these tools represent a useful addition to our existing library of genetic research tools (Figure 1).
Figure 1.
SNP query tool snapshot highlighting SNP filtering. Multiple filtering options include: validation (T/F), HapMap (31) (T/F), Location, avHet, avHetSE, HapMap Frequency (CEU, CHB, JPT, YRI), SIFT score and UCSC conservation. Users can preview current filtering criteria by scrolling over pop-up window link. Once SNPs are selected, Haploview like images can be rendered showing HapMap LD structure (lower right).
METHODS AND USAGE
Web interface and data organization
The SNP and mutation data is parsed directly from Swiss-Prot (currently build 51.0) and dbSNP (currently build 126) without curation, other than to remove any annotations that do not map to the wild-type amino acid in the referenced sequence. The gene model provided by MutDB is organized, using gene information extracted from a local copy of the UCSC Human Genome Annotation Database (ver. Hg18, http://genome.ucsc.edu/) (25). We also use Ensembl (ver. 41_36c, http://www.ensembl.org/) (26) for some gene name cross-references. We attempt to keep pages organized by Entrez Gene ID with the most representative transcript as the primary gene page. Other known mRNA transcripts annotated in the UCSC Genome Annotation Database are listed at the bottom of the page with their annotations. This data may be browsed alphabetically by gene symbol or by employing one of several search methods, including keyword, gene symbol, protein or Refseq ID, and individual variant identifier. Each gene is given its own page for display, providing a list of related SNPs and mutations classified by their effects on the protein product, as well as a pictorial representation of the sequence including points of conservation, location of exons and location of variants. Links to corresponding Swiss-Prot and dbSNP pages, a short description of the gene, and the related chromosome name are supplied.
Each variant is annotated within its own page providing further details, which includes the protein sequence, if known, and any related Protein Data Bank (PDB) (http://www.rcsb.org/pdb/) (27) structures, KEGG Pathways, HapMap data and Entrez Gene information. We describe important aspects of our annotation pipeline below.
Protein structure annotations
Protein structural mapping for each amino acid substitution is performed by aligning the query sequence to each high scoring segment pair (HSP) from BLAST (28) search results using BioPerl scripts (29). BLAST results used for alignment come only from PDB (using a sequence data file downloaded in January of 2007) and are limited to those with 100% identity to the original sequence. These pairwise alignments are then used to map wild-type and mutation sequence to structure sequence. The annotated mutations that are mapped to a structure can be displayed using the integrated Jmol visualization tool (http://jmol.sourceforge.net/) or in extensions developed for UCSF Chimera (30) and Delano Scientific PyMOL (http://pymol.sourceforge.net/). To download the extensions visit http://lifescienceweb.org/.
Function annotations
We provide links to other tools that provide predictions of functional or molecular disruptions caused by an amino acid substitution. These include SNPs3D (5), PolyPhen (4), SIFT (3), PolyDoms (9), PMUT (7) and PANTHER (6) and are deep linked directly to the gene or SNP page, if available. Sorting Intolerant from Tolerant (SIFT) scores (3) and their associated predictions are supplied for each variant causing an amino acid substitution. Variants with low confidence scores are marked with an asterisk. Here, again, the source Swiss-Prot and dbSNP pages are linked.
Visualization on KEGG pathways
We have augmented MutDB annotations with KEGG pathways using KEGG web services (23). This enables visualizing proteins, mutations and pathways on approximately 188 human pathways found in KEGG. The addition of a link, ‘Visualize Pathways’, on the MutDB gene page takes the user to a page listing the names of all KEGG pathways involving the gene. When a pathway is chosen, the user is taken to a new page displaying the pathway and a list of involved genes and their associated phenotypes.
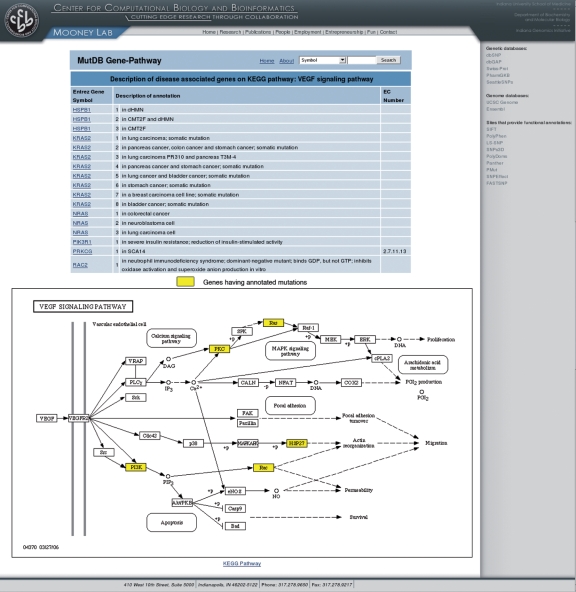
All genes containing a SNP denoted as having a disease annotation or comment (per Swiss-Prot) are colored yellow in the pathway. This page is also hyper-linked to KEGG and MutDB. This functionality makes use of KEGG SOAP-based web services with supplementary data saved locally (Figure 2).
Figure 2.
MutDB-KEGG integration example of the VEGF Pathway. This pathway shows all proteins with SNPs or Swiss-Prot mutations and all unique diseases and comments provided by Swiss-Prot (top). The VEGF signaling pathway showing proteins with mutations in yellow (bottom).
SNP query tool
A recent addition to our toolset is a SNP query tool that enables querying and exporting sets of SNPs that share selected features. The SNP query tool requires two sequence-tagged site (STS) markers or dbSNP reference cluster IDs (rs#) as input and returns all SNPs between the markers. The tool uses AJAX and a paging scheme to increase responsiveness upon large data sets. AJAX enhances speed by exchanging small amounts of data with the server, so the entire web page need not be reloaded each time the user makes a change. This technique along with the broad filtering options provide for an interactive tool.
Users may filter SNPs by manual selection or one of the filtering criteria. There are currently eleven filter options: validation status in dbSNP, hapmap status, location (functional class), avHet (average heterozygosity in dbSNP), avHetSE (SE for the average heterozygosity in dbSNP), CEU (CEPH—Utah residents with ancestry from northern and western Europe frequencies in HapMap), CHB (Han Chinese in Beijing, China), JPT (Japanese in Tokyo, Japan), YRI (Yoruba in Ibadan, Nigeria), SIFT score (3) and conservation score [based on the UCSC Genome Annotation Database conservation (25)]. The conservation score is the averaged 10-mer window of conservation values around each SNP derived from alignments of the 16 vertebrate species in the UCSC Annotated Genome Database.
A user can authenticate to enter the tool or visit as a guest, and may save each session and return later. Retrieval of sequence surrounding the SNP and exportation of SNP data to Microsoft Excel are easily performed via provided links. Excel output includes the dbSNP rsID, primer sequences, and the polymorphic alleles. The tool displays a PNG image containing RefSeq transcript information and location information for all selected SNPs indexed by function type using the UCSC Genome Annotation Database. A user may also visualize linkage disequilibrium for up to 200 selected SNPs in a Haploview (24) like structure. The SNP query tool is located at http://www.mutdb.org/snp and is linked from each page (Figure 1).
Continued web services support
MutDB continues to support its SOAP-based web services. The web services can be accessed via http://www.lifescienceweb.org. This interface is used to communicate to the structural visualization extensions for UCSF Chimera and Delano Scientific PyMOL.
Most accessed gene pages
In MutDB, the most accessed genes may give insight into the current interests of researchers. The most accessed genes from October 2005 to January 2007 are listed in Table 1. Not surprisingly, the most accessed genes also have many mutations associated with them and are what we would consider to be well-studied disease-associated genes.
Table 1.
Top 15 accessed genes on MutDB from October 2005 to January 2007
| Symbol | Name |
|---|---|
| 1. BRCA1 | Breast cancer 1, early onset |
| 2. CFTR | Cystic fibrosis transmembrane conductance |
| 3. AR | Androgen receptor |
| 4. APOE | Apolipoprotein E precursor |
| 5. ATP7B | ATPase, Cu++transporting, beta polypeptide |
| 6. TP53 | Tumor protein p53 |
| 7. CD53 | CD53 antigen |
| 8. BRCA2 | Breast cancer 2, early onset |
| 9. FBN1 | Fibrillin 1 |
| 10. APC | Adenomatosis polyposis coli |
| 11. NOTCH3 | Notch homolog 3 |
| 12. KALRN | Kalirin, RhoGEF kinase |
| 13. CYP2D6 | Cytochrome P450, family 2, subfamily D |
| 14. RET | Ret proto-oncogene |
| 15. HBB | Beta globin |
BRCA1, CFTR, AR and APOE are the most requested pages within MutDB.
Future
Understanding the underlying molecular causes of disease remains an important area for research. We continue to investigate annotations that are useful for hypothesis generation and directing experimental validation. While we continue to update the database as new annotations become available, we are also adding useful annotations outside of protein amino acid changes such as noncoding, synonymous and intronic variation.
ACKNOWLEDGEMENTS
We would like to thank Shoji Ichikawa and Somying Promso for helpful comments on the SNP query tool. We are supported by NLM K22LM009135 (PI: Mooney), P01AG018397 (PI: Econs), a grant from IU Biomedical Research Council, an RSFG grant from IUPUI, the Showalter Trust and the Indiana Genomics Initiative. The Indiana Genomics Initiative (INGEN) is supported in part by the Lilly Endowment. RH is supported by Indiana Pervasive Computing Research (IPCRES) Initiative. Funding to pay the Open Access publication charges for this article was provided by NLM K22LM009135 (PI: Mooney).
Conflict of interest statement. None declared.
REFERENCES
- 1.Mooney S. Bioinformatics approaches and resources for single nucleotide polymorphism functional analysis. Brief Bioinform. 2005;6:44–56. doi: 10.1093/bib/6.1.44. [DOI] [PubMed] [Google Scholar]
- 2.Ng PC, Henikoff S. Predicting the effects of amino Acid substitutions on protein function. Annu. Rev. Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 3.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sunyaev S, Ramensky V, Koch I, Lathe W., III, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum. Mol. Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 5.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrer-Costa C, Gelpi JL, Zamakola L, Parraga I, de la Cruz X, Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 8.Karchin R, Diekhans M, Kelly L, Thomas DJ, Pieper U, Eswar N, Haussler D, Sali A. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 9.Jegga AG, Gowrisankar S, Chen J, Aronow BJ. PolyDoms: a whole genome database for the identification of non-synonymous coding SNPs with the potential to impact disease. Nucleic Acids Res. 2007;35:D700–D706. doi: 10.1093/nar/gkl826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reumers J, Schymkowitz J, Ferkinghoff-Borg J, Stricher F, Serrano L, Rousseau F. SNPeffect: a database mapping molecular phenotypic effects of human non-synonymous coding SNPs. Nucleic Acids Res. 2005;33:D527–D532. doi: 10.1093/nar/gki086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saunders CT, Baker D. Evaluation of structural and evolutionary contributions to deleterious mutation prediction. J. Mol. Biol. 2002;322:891–901. doi: 10.1016/s0022-2836(02)00813-6. [DOI] [PubMed] [Google Scholar]
- 13.Cavallo A, Martin AC. Mapping SNPs to protein sequence and structure data. Bioinformatics (Oxford, England) 2005;21:1443–1450. doi: 10.1093/bioinformatics/bti220. [DOI] [PubMed] [Google Scholar]
- 14.Chasman D, Adams RM. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J. Mol. Biol. 2001;307:683–706. doi: 10.1006/jmbi.2001.4510. [DOI] [PubMed] [Google Scholar]
- 15.Dobson RJ, Munroe PB, Caulfield MJ, Saqi MA. Predicting deleterious nsSNPs: an analysis of sequence and structural attributes. BMC bioinformatics. 2006;7:217. doi: 10.1186/1471-2105-7-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnan VG, Westhead DR. A comparative study of machine-learning methods to predict the effects of single nucleotide polymorphisms on protein function. Bioinformatics (Oxford, England) 2003;19:2199–02209. doi: 10.1093/bioinformatics/btg297. [DOI] [PubMed] [Google Scholar]
- 17.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mooney SD, Altman RB. MutDB: annotating human variation with functionally relevant data. Bioinformatics. 2003;19:1858–1860. doi: 10.1093/bioinformatics/btg241. [DOI] [PubMed] [Google Scholar]
- 19.Dantzer J, Moad C, Heiland R, Mooney S. MutDB services: interactive structural analysis of mutation data. Nucleic Acids Res. 2005;33:W311–W314. doi: 10.1093/nar/gki404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheeler DL, Church DM, Edgar R, Federhen S, Helmberg W, Madden TL, Pontius JU, Schuler GD, Schriml LM, et al. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 2004;32(Database issue):D35–D40. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 25.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: A New Generation Of Protein Database Search Tools. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]