Abstract
PlantGDB (http://www.plantgdb.org/) is a genomics database encompassing sequence data for green plants (Viridiplantae). PlantGDB provides annotated transcript assemblies for >100 plant species, with transcripts mapped to their cognate genomic context where available, integrated with a variety of sequence analysis tools and web services. For 14 plant species with emerging or complete genome sequence, PlantGDB's genome browsers (xGDB) serve as a graphical interface for viewing, evaluating and annotating transcript and protein alignments to chromosome or bacterial artificial chromosome (BAC)-based genome assemblies. Annotation is facilitated by the integrated yrGATE module for community curation of gene models. Novel web services at PlantGDB include Tracembler, an iterative alignment tool that generates contigs from GenBank trace file data and BioExtract Server, a web-based server for executing custom sequence analysis workflows. PlantGDB also hosts a plant genomics research outreach portal (PGROP) that facilitates access to a large number of resources for research and training.
INTRODUCTION
PlantGDB serves the plant research community by providing access to plant sequence data as well as a variety of sequence and genome analysis tools in a single online resource [(1,2); Table 1]. This update outlines recent developments at PlantGDB that have expanded its usefulness as a tool for comparative genomics. Key features include: expanded EST assemblies; new genome browsers for a larger number of species; overnight annotation of emerging genome sequences; and novel tools for sequence retrieval and analysis, including an innovative system for the creation and management of workflows that integrates database queries, linked web services, and local tools.
Table 1.
Sequence resources and analytical tools available at PlantGDB
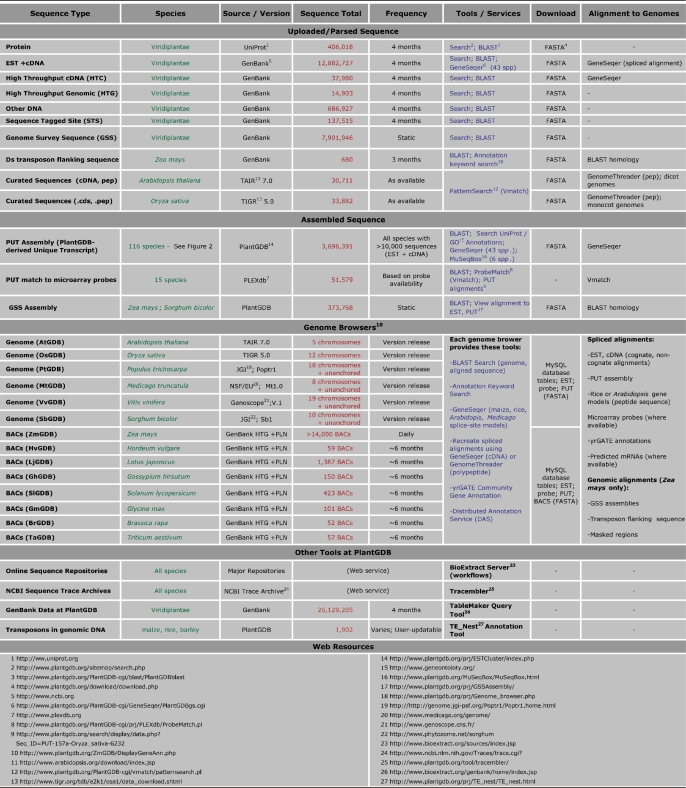 |
In this table, sequence resources are divided into four categories: Uploaded Sequence, Assembled Sequence, Genome Browsers, and Other Tools. For each resource in column 1, the species available, source/version, current sequence count, update frequency, tool/services, download options, and alignment to genome are shown in adjacent cells. Web links for both external and internal data/tool sources are indicated with superscript numbers and are listed at the end of the table under Web Resources. Sequence counts displayed here are as of 30 October 2007.
DATABASE FEATURES AND ADDITIONS
Plant sequence data and transcript assemblies
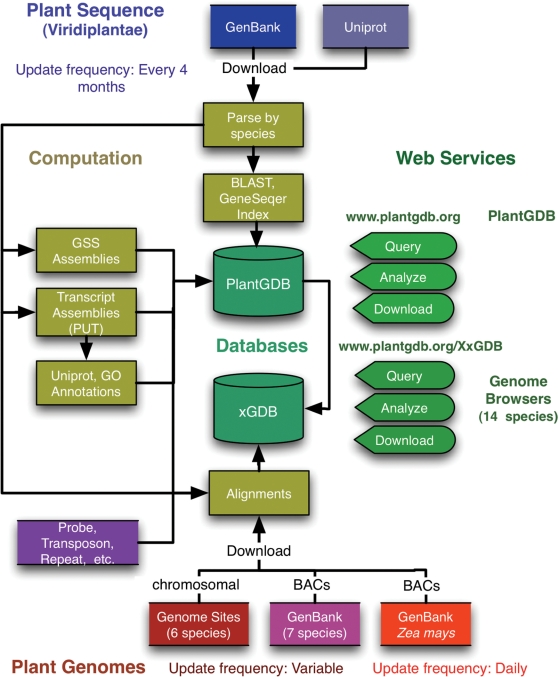
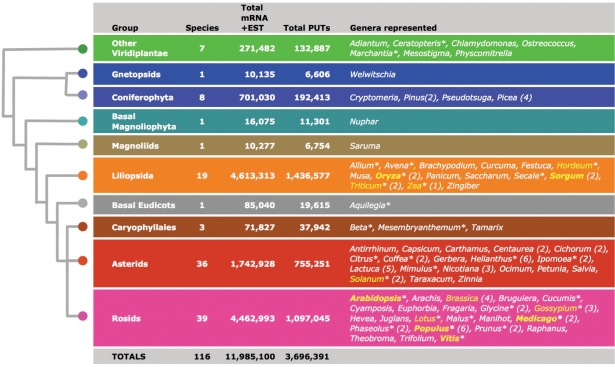
PlantGDB periodically uploads and parses all Viridiplantae sequences from GenBank (3) and Uniprot (4) into ∼70 000 individual data sets according to species or subspecies origin (Figure 1). PlantGDB's sequence data are refreshed approximately every 4 months, coinciding with alternate bimonthly GenBank version releases. GenBank and Uniprot sequences are uploaded, parsed by (sub)species, indexed for BLAST (5) and GeneSeqer (6) analysis, and loaded into MySQL tables. For all species with >10 000 published transcripts, a non-redundant set of PlantGDB-generated Unique Transcripts (PUTs) is generated using a custom assembly pipeline (http://www.plantgdb.org/prj/ESTCluster/PUT_procedure.php). PUTs are aligned to UniProt entries using BLAST, and the best matches (if any) and UniProt-associated Gene Ontology (GO) annotations (7) are stored. Currently, 116 species have PUT assemblies at PlantGDB, spanning diverse taxonomic groups (Figure 2). Users can track assembly progress in PlantGDB at http://www.plantgdb.org/prj/ESTCluster/progress.php. PlantGDB also provides genome survey sequence (GSS) assemblies for maize and sorghum (http://www.plantgdb.org/prj/GSSAssembly/). All PlantGDB sequence data (raw and processed) are available for download in a variety of file formats at http://www.plantgdb.org/download/download.php. In addition, all Zea mays sequence data at PlantGDB are uploaded monthly to MaizeGDB, the central repository for maize genetic information (http://www.maizegdb.org) (8).
Figure 1.
Database schema for PlantGDB, showing data sources, update frequency, computation and web services. PlantGDB is accessible at http://www.plantgdb.org, and genome browsers are accessible at http://www.plantgdb.org/XxGDB, where Xx is the first letter of the genus and species (e.g. AtGDB = Arabidopsis thaliana genome database).
Figure 2.
Transcript assemblies (PUTs) at PlantGDB, grouped by taxonomic affiliation. Sequence totals displayed here are as of 30 October 2007. Parentheses indicate the number of species/subspecies per genus. Genera highlighted in yellow are associated with a genome browser at PlantGDB; an underscore indicates chromosome-based genome browsers. An asterisk designates genera for which PlantGDB provides preprocessed GeneSeqer indices for quick access to spliced alignments.
Query and analysis tools
Data housed at PlantGDB are stored in MySQL tables and can be queried by accession number, GI number or text search. TableMaker, a search module for querying and retrieving PlantGDB's GenBank data in tabular format, described previously (1), has been expanded to include a new query wizard to simplify the search process for users not familiar with GenBank data models (http://www.bioextract.org/genbank/home/index.jsp).
For sequence similarity searching, a batch NCBI-BLAST tool is available for querying any combination of plant species data sets against up to 100 query sequences at a time (http://www.plantgdb.org/PlantGDB-cgi/blast/PlantGDBblast). For specialized queries, PatternSearch (http://www.plantgdb.org/PlantGDB-cgi/vmatch/patternsearch.pl) interrogates the database for relatively short matches possibly interspersed with mismatches and indels, and ProbeMatch (http://www.plantdb.org/PlantGDB-cgi/prj/PLEXdb/ProbeMatch.pl) allows users to match sequence to array probes and link to array probe databases.
PlantGDB also provides online access to GeneSeqer alignment software, allowing the user to calculate spliced alignments of expressed transcripts to a target genomic sequence, as described previously (1) (http://www.plantgdb.org/PlantGDB-cgi/GeneSeqer/PlantGDBgs.cgi). Currently, transcript data sets (EST, cDNA, PUT) from 50 species are preprocessed at PlantGDB to allow rapid online GeneSeqer analysis, and a range of splice site models and alignment parameters can be specified at runtime.
PlantGDB provides sequence analysis tools that automate processes that normally require iterative searching or tedious parsing of information. Tracembler (http://www.plantgdb.org/tool/tracembler/) allows the user to do chromosome walks with pre-assembly trace data by performing an iterated search of NCBI's trace archives with a seed sequence (9). Tracembler invokes CAP3 (10) to assemble a contig sequence from one or more automated rounds of BLAST analysis and displays a pairwise alignment between contig and seed sequence. MuSeqBox (http://www.plantgdb.org/MuSeqBox/MuSeqBox.html) is an online tool for generating tabular output from multiple BLAST queries, based on user-specified thresholds (11). The tool also contains algorithms for detecting potential alternate splicing and full-length transcripts. MuSeqBox provides pre-computed Uniprot BLASTx data sets for maize, sorghum, barley, Arabidopsis and rice PUTs, allowing the user to generate filtered, tabulated output. Alternatively, the user can upload a custom BLAST output file.
Bioinformatics workflow tools
A bioinformatics research project may utilize a variety of query and computational tools, some web-based and others local to the user, which may be carried out in serial fashion along with parsing and formatting for input or display. There is a growing need for systems that can integrate disparate tools and workflows in a way that automates the process of input/output, computation and documentation. The PlantGDB-associated BioExtract Server (http://www.bioextract.org/login/Login.html) addresses this need by providing researchers with a web interface that allows them to query sequence databases, analyze data with web-based as well as local bioinformatics tools, save results and create and manage workflows using a directed acyclic graph (DAG) model (Lushbough,C., Bergman,M.K., Lawrence,C.J., Jennewein,D. and Brendel,V. BioExtract server – an integrated system to access and analyze heterogeneous, distributed biomolecular data. Submitted for publication.). As a simple example, a user could develop a workflow that performs a BLAST search, retrieves peptide sequences from query results, eliminates redundant sequences and produces a multiple sequence alignment output. BioExtract workflows can be paused, modified, saved, shared with an online workgroup or the world, and documented electronically for future reference.
Genome browsers
PlantGDB provides genome browsers (xGDB) for 14 plant species whose genomes have been completely or partially sequenced (12). AtGDB, OsGDB, MtGDB, SbGDB, VvGDB and PtGDB are chromosome-based genome browsers for Arabidopsis thaliana (thale cress), Oryza sativa (rice), Medicago truncatula (barrel medic), Sorghum bicolor (sorghum), Vitis vinifera (grapevine) and Populus trichocarpa (western balsam poplar), respectively, while ZmGDB, GmGDB, HvGDB, SlGDB, GhGDB, TaGDB, BrGDB and LjGDB are BAC-based browsers for Zea mays (corn or maize), Glycine max (soybean), Hordeum vulgare (barley), Solanum lycopersicum (tomato), Gossypium hirsutum (cotton), Triticum aestivum (bread wheat), Brassica rapa (field mustard) and Lotus japonicus, respectively (http://www.plantgdb.org/prj/Genome_browser.php).
The xGDB browser Context View (Figure 3B) displays current gene model annotation together with high quality, cognate and non-cognate GeneSeqer alignments of ESTs, cDNAs, and PUTs to genomic sequence. Similarly, Oryza sativa predicted polypeptides from TIGR (http://rice.tigr.org/tdb/e2k1/osa1/) (13) and/or Arabidopsis thaliana predicted polypeptides from TAIR (http://www.arabidopsis.org/) (14) are splice-aligned to genomic sequence using GenomeThreader (15) and displayed in the same window. For species with microarray probe sequence, these are downloaded from the microarray database at PLEXdb (16), aligned relative to PUT assemblies and displayed. Significantly, users can view spliced alignments in a genomic region at the nucleotide level and also retrieve quality scores and provenance information for any spliced alignment displayed at xGDB. Additional sequence alignments, including GSS contigs and repeat masked regions, are displayed for some genomes. A subset of xGDB annotation data are accessible through the Distributed Annotation System (DAS) (http://www.biodas.org/). These data can be downloaded for further analysis, or alternatively imported into another genome browser capable of importing DAS formatted data.
Figure 3.
Screenshots from ZmGDB and yrGATE illustrate the use of online tools for gene discovery and community gene annotation. (A) A web-accessible table of Z. mays BACs (alternately shaded) displaying (left to right) the BAC GI, BAC clone name, followed by the ID, start/end coordinates and functional annotation of splice-aligned TIGR-predicted proteins from O. sativa and finally the ZmGDB entry date. All fields are searchable and each row is linked via column 1 to a genome browser view of the BAC region. This table is currently updated daily at ZmGDB (http://www.plantgdb.org/ZmGDB/DisplayGeneAnn.php). (Similar tables are available for eight other BAC-based xGDB browsers.) Note that a region of BAC GI 156523432 is aligned to three paralogous rice predicted polypeptides, annotated as ‘autophagy-related protein 8 precursor’. Clicking on the BAC GI ‘156523432’ in table column 1 (circled) brings up a BAC/Clone Context View of the specified region (B), showing spliced alignments to the rice predicted polypeptides (black), along with other alignment data, in this case maize cDNAs (blue) and maize ESTs (red). Note the evidence for alternative splicing among the maize ESTs (circles) suggesting at least two alternate transcripts (labeled 1 and 2). The user has the option to explore and annotate this variation using yrGATE. (C) Launching the yrGATE annotation tool displays scrolling list of evidence scores and supporting exons for all exon coordinates at a locus (alternative splice coordinates for 1 and 2 are circled). The user can build a complete gene model on screen by selecting each desired exon and then compare the resulting open reading frame to known proteins using BLAST (data not shown). (D) The chosen gene model is displayed graphically and will be published on the ZmGDB browser following curation by PlantGDB staff. Shown here are yrGATE models for the two putative splice variants, with translation start/stop positions indicated by triangles. (E) Predicted protein sequence for the two yrGATE gene models. This example illustrates how xGDB and yrGATE can be used to identify and publish gene model predictions quickly and easily, enhancing the community genome knowledge base for maize as well as facilitating hypothesis-driven research.
Key design features of the xGDB browsers are modularity and expandability, such that a browser for a newly emerging genome can be rapidly deployed and populated with computed alignments. For the user, xGDB provides a clean, easy to navigate interface with links to internal and external data sources, zoomable and customizable views of transcript alignments, BLAST, search tools and the ability to evaluate alignments and contribute new annotations online (see next section). The browsers are designed with Perl/PHP and MySQL database management, and complete code is available from SourceForge (http://xgdb.sourceforge.net/) for standalone installations.
Community annotation
Although excellent tools are available for defining genic regions and variant transcript forms from evidence-based data as well as ab initio prediction, models can often be improved further by human curated annotation. PlantGDB's yrGATE (http://www.plantgdb.org/prj/yrGATE/) is a recently developed tool for community annotation of gene models that is integrated with PlantGDB's xGDB genome browsers (17). From a single browser window the user can rapidly evaluate a selected region for intron/exon structures based on any combination of EST/cDNA evidence and ab initio prediction, compare the model with known proteins via GenBank BLAST, and submit the annotation for review and publication on the genome browser. To assist in the identification of gene models in need of annotation, the Genome Annotation Evaluation (GAEVAL) module generates quality scores for gene structure predictions and classifies cases of incongruence of the annotation with experimental evidence (http://www.plantgdb.org/AtGDB-chtml/gaeval/). The yrGATE tool is available for both BAC and chromosome-based xGDB browsers and is being used to communicate evidence-based gene models to the A. thaliana genome database, TAIR (18). Figure 1 shows an example of how yrGATE can be used, together with xGDB's annotation tables and genome browser, to identify and annotate potential splicing variants for a gene of interest in maize.
Pipelines for genome annotation
Genome browsers at PlantGDB are refreshed on a timetable that depends on the pace of accumulation of new genomic or transcript sequence data or assemblies for the respective species. New spliced alignments are calculated for ESTs, cDNAs and PUTs as well as for other sequence types (where available) and data are uploaded. To match the rapid pace with which some genomes are being sequenced, PlantGDB staff have developed and implemented an automated genome data pipeline for species with rapidly expanding sequence data, using Zea mays as an initial example. In 2007, new maize BAC sequences began to be deposited in GenBank at the rate of over 60 BACs or ∼10 Mb of sequence per day (http://www.maizesequence.org). In addition, there is a growing catalog of transposable element-tagged maize genomic sequence in GenBank, facilitating reverse genetics in maize (http://www.plantgdb.org/prj/AcDsTagging/) (19) as well as a large repository of EST sequence-derived PUTs. PlantGDB's daily Z. mays pipeline downloads and processes all new maize BACs with transcript, protein, microarray probe, transposon insertion tag and other genomic alignments, and displays the cumulative output for all BACs in ZmGDB (the xGDB browser for Z. mays; http://www.plantgdb.org/ZmGDB/) within 12 h. The pipeline also updates BLAST and sequence download resources daily. Significantly, the pipeline also generates a browsable, searchable, tabular output of rice gene models and putatively transposon-tagged genes for the entire BAC data set (http://www.plantgdb.org/ZmGDB/DisplayGeneAnn.php), providing a powerful and timely gene discovery tool for researchers (Figure 3). This effort represents an early implementation of a real-time, high-throughput, discovery-oriented annotation process using automated workflows.
Outreach
The Plant Genome Outreach Portal (PGROP) at PlantGDB provides a portal for plant genomics resources online as well as a repository of outreach content, serving the needs of a wide-ranging audience from high school through postgraduate (20). Users can query for resources or add a resource using simple online tools, and query results are ranked via algorithms that highlight the most popular resources.
Future directions
PlantGDB will continue to provide comprehensive and up-to-date plant sequence information online and available for download. As additional genomes become available, xGDB browsers will be expanded, with additional annotations contemplated for certain species [e.g. tracks for transcription factor binding sites and conserved non-coding sequences (21)]. Also planned are additional comparative genomics tools such as SynBrowse (22), expanded DAS import and export, and the development of qualitative (e.g. quality score) and quantitative (e.g. library) filters for spliced alignments. Expanded help and tutorial sections are also under development.
CONCLUSIONS
PlantGDB has expanded greatly in scope since 2004, providing today a wide range of data sets, query methods and analysis tools for researchers interested in comparative plant genomics or gene discovery research. The site aims to complement other, more specialized plant genome sites by providing comprehensive plant sequence data as well as a suite of tools and genome browsers that emphasize spliced alignment of cognate and non-cognate transcripts and similar protein sequences. PlantGDB also addresses the need for timely access to, and processing of, high-volume informatics data through use of automated daily data pipelines (e.g. maize BAC pipeline) and online workflow tools (e.g. BioExtract Server and Tracembler). With the yrGATE community annotation tool, PlantGDB facilitates the sharing of user-generated gene annotation information across the entire plant research community.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Q. Dong, Dr S.D. Schlueter and Dr B.-B.Wang for their many contributions to PlantGDB over the years, M. Brekke for system support, and the many PlantGDB users who have helped us to improve this resource by providing feedback and suggestions. This work is supported in part by a grant from the National Science Foundation Plant Genome Research Program to V.B., C.J.L, and C.L. (DBI-0606909). Funding to pay the Open Access publication charges for this article was provided by the cited NSF grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dong Q, Schlueter SD, Brendel V. PlantGDB, plant genome database and analysis tools. Nucleic Acids Res. 2004;32:D354–D359. doi: 10.1093/nar/gkh046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong Q, Lawrence CJ, Schlueter SD, Wilkerson MD, Kurtz S, Lushbough C, Brendel V. Comparative plant genomics resources at PlantGDB. Plant Physiol. 2005;139:610–618. doi: 10.1104/pp.104.059212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bensen DA, Karsch-Mizrachi I, Lipman J, Wheeler DL. GenBank. Nucleic Acids Res. 2006;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Brendel V, Xing L, Zhu W. Gene structure prediction from consensus spliced alignment of multiple ESTs matching the same genomic locus. Bioinformatics. 2004;20:1157–1169. doi: 10.1093/bioinformatics/bth058. [DOI] [PubMed] [Google Scholar]
- 7.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence CJ, Schaeffer ML, Seigfried TE, Campbell DA, Harper LC. MaizeGDB's new data types, resources and activities. Nucleic Acids Res. 2007;35:D895–D900. doi: 10.1093/nar/gkl1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong Q, Wilkerson MD, Brendel V. Tracembler - software for in silico chromosome walking in unassembled genomes. BMC Bioinformatics. 2007;8:151. doi: 10.1186/1471-2105-8-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing L, Brendel V. Multi-query sequence BLAST output examination with MuSeqBox. Bioinformatics Not. 2001;17:744–745. doi: 10.1093/bioinformatics/17.8.744. [DOI] [PubMed] [Google Scholar]
- 12.Schlueter SD, Wilkerson MD, Dong Q, Brendel V. xGDB: open-source computational infrastructure for the integrated evaluation and analysis of genome features. Genome Biol. 2006;7:R111. doi: 10.1186/gb-2006-7-11-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gremme G, Brendel V, Sparks ME, Kurtz S. Engineering a software tool for gene structure prediction in higher organisms. Info. Sof. Technol. 2005;47:965–978. [Google Scholar]
- 16.Shen L, Gong J, Caldo RA, Nettleton D, Cook D, Wise RP, Dickerson JA. BarleyBase — An expression profiling database for plant genomics. Nucleic Acids Res. 2005;33:D614–D618. doi: 10.1093/nar/gki123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkerson MD, Schlueter SD, Brendel V. yrGATE: a web-based gene-structure annotation tool for the identification and dissemination of eukaryotic genes. Genome Biol. 2006;7:R58. doi: 10.1186/gb-2006-7-7-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlueter SD, Wilkerson MD, Huala E, Rhee SY, Brendel V. Community-based gene structure annotation for the Arabidopsis thaliana genome. Trends Plant Sci. 2005;10:9–14. doi: 10.1016/j.tplants.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Conrad LJ, Brutnell TP. Ac-Immobilized, a stable source of activator transposase that mediates sporophytic and gametophytic excision of dissociation elements in maize. Genetics. 2005;171:1999–2012. doi: 10.1534/genetics.105.046623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baran S, Lawrence CJ, Brendel V. PGROP - A gateway to plant genome research ‘outreach’ programs and activities. Plant Physiol. 2004;134:889. doi: 10.1104/pp.103.038414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas BC, Rapaka L, Lyons E, Pedersen B, Freeling M. Arabidopsis intragenomic conserved noncoding sequence. Proc. Natl Acad. Sci. USA. 2007;104:3348–3353. doi: 10.1073/pnas.0611574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan X, Stein L, Brendel V. SynBrowse: a synteny browser for comparative sequence analysis. Bioinformatics. 2005;21:3461–3468. doi: 10.1093/bioinformatics/bti555. [DOI] [PubMed] [Google Scholar]