Abstract
Y chromosome deletions arise frequently in human populations, where they cause sex reversal and Turner syndrome and predispose individuals to infertility and germ cell cancer. Knowledge of the nucleotide sequence of the male-specific region of the Y chromosome (MSY) makes it possible to precisely demarcate such deletions and the repertoires of genes lost, offering insights into mechanisms of deletion and the molecular etiologies of associated phenotypes. Such deletion mapping is usually conducted using polymerase chain reaction (PCR) assays for the presence or absence of a series of Y-chromosomal DNA markers, or sequence-tagged sites (STSs). In the course of mapping intact and aberrant Y chromosomes during the past two decades, we and our colleagues have developed robust PCR assays for 1287 Y-specific STSs. These PCR assays amplify 1698 loci at an average spacing of <14 kb across the MSY euchromatin. To facilitate mapping of deletions, we have compiled a database of these STSs, MSY Breakpoint Mapper (http://breakpointmapper.wi.mit.edu/). When queried, this online database provides regionally targeted catalogs of STSs and nearby genes. MSY Breakpoint Mapper is useful for efficiently and systematically defining the breakpoint(s) of virtually any naturally occurring Y chromosome deletion.
INTRODUCTION
Most boys and men carry the entirety of the Y chromosome, including the male-specific region (MSY) (1), while most girls and women carry none of it. However, some males and some females possess a portion but not all of the MSY. This is commonly the result of a translocation involving the Y chromosome and a second chromosome, or of an interstitial or other deletion within the Y chromosome (2–30). Taken together, these naturally occurring deletions of the MSY are abundant, and they are diverse in structure and origin.
In recent decades, studies of these MSY deletions have propelled major advances in our understanding of Y chromosome function and structure. These advances have included the elucidation of sex reversal syndromes (e.g. XX males and XY females) (4,5,8–13,15,21), the identification of the sex-determining gene SRY (31), the emergence of MSY deletions as the most common known genetic cause of spermatogenic failure (18,20,22–27) and the implication of MSY genes in the etiology of gonadoblastoma and testis cancer (19,32,33). The first comprehensive maps of the human Y chromosome were based on naturally occurring deletions: Y-specific restriction fragments and, subsequently, Y-specific sequence-tagged sites (STSs) were ordered by testing for their presence or absence in genomic DNAs of series of individuals with partial Y chromosomes (6,7,14). In turn, these deletion-mapped STSs were used to build scaffolds of overlapping recombinant DNA clones (34), ultimately leading to a tiling path of BAC clones for the sequencing of the MSY (1,35).
With the availability of the nucleotide sequence of the MSY (1), we and other investigators have employed a growing catalog of STSs in an expanded effort to identify MSY deletions and to understand their phenotypic effects and modes of origin. For example, several recurrent interstitial deletions associated with spermatogenic failure have recently been defined at the DNA-sequence level (22–27). Such studies have underscored the roles of many MSY genes in reproduction and have identified ectopic homologous recombination as a prominent mechanism of MSY deletion.
Although a few such classes of MSY deletions have been examined at the DNA-sequence level, many others have yet to be thoroughly explored. These include translocations of the Y chromosome to the X chromosome or to an autosome, Y isochromosomes and ring chromosomes (2,14,16,17). Such structural abnormalities are associated with a wide array of phenotypes, including spermatogenic failure, manifestations of Turner syndrome, ambiguous external genitalia, gonadoblastoma and delay in development and growth.
Detailed DNA-sequence analysis of uncharacterized MSY deletions is most readily accomplished with Y-specific STS assays, which employ the polymerase chain reaction (PCR). Each such STS assay provides a straightforward means of determining the presence or absence, in a sample of human genomic DNA, of a specific point along the length of the Y chromosome (14). In the course of analysing normal and aberrant Y chromosomes over the past two decades, we and our colleagues have generated 1287 robust, Y-specific STSs. Here we present MSY Breakpoint Mapper (http://breakpointmapper.wi.mit.edu/), a database of these STSs and an interface for use in examining MSY deletions.
DATABASE OF STSs
The MSY Breakpoint Mapper database contains 1287 Y-specific STSs generated and tested during the past two decades (1,14,18,22,26,35,36). Among these are 695 previously unreported STSs that our laboratory has recently deposited in GenBank. Each of the 1287 archived STSs is operationally defined by a PCR assay. In most cases, PCR primers were selected using Primer3 (37). Each of the 1287 PCR assays was empirically demonstrated to yield the expected product when normal male genomic DNAs were employed as templates, but not when normal female genomic DNAs were employed. Of these 1287 PCR assays, 1277 amplify single-copy or low-copy-number sequences found in the euchromatic portions of the MSY. The remaining 10 PCR assays amplify highly repetitive heterochromatic sequences found only in the Y chromosome (Table 1).
Table 1.
Numbers of PCR assays (STSs) and corresponding amplified loci archived in the database
| Sequence type | Number of PCR assays (STSs) | Number of loci amplified | |||||
|---|---|---|---|---|---|---|---|
| Single-copy | Multi-copy | Total | Single-copy | Multi-copy | Total | Density | |
| Euchromatina | 992 | 285 | 1277 | 992 | 706 | 1698 | 1 per 13.6 kbc |
| Heterochromatinb | NA | NA | 10 | NA | NA | NA | NA |
aSTSs that span euchromatin–heterochromatin boundaries are categorized here as deriving from euchromatin.
bDue to the repetitive nature of MSY heterochromatic sequences, it is not possible to determine the copy number of STSs that derive from such sequences or the total number of loci amplified.
cCalculation based on the MSYs 23 Mb of euchromatic sequence.
For 992 of the 1277 euchromatic STSs, the PCR assay amplifies a single, unique site in the MSY. Each of these 992 single-copy STSs is therefore useful in assaying the presence or absence of a unique point along the length of the Y chromosome. It is noteworthy that, for several dozen of these single-copy STSs, the unique target site exhibits sequence similarity to one or more other points along the Y chromosome. For each of these STSs, the corresponding PCR assay is nonetheless specific to the targeted locus, as shown using negative control templates of two kinds: (i) genomic DNAs from individuals with Y chromosomes known to be deleted for the STS target site, but also known to carry the sequence-similar locus and (ii) BAC clones lacking the STS target site, but carrying the sequence-similar locus. In each case, the PCR assay proved to be specific to the STS target site.
The remaining 285 (of 1277) euchromatic STSs are not single-copy but multi-copy. The MSY contains many lengthy, dispersed repeats or ‘amplicons’ that exhibit >99.9% sequence identity. A PCR assay for such sequences will necessarily amplify multiple loci. For each of the 285 multi-copy STSs, the PCR assay amplifies identical or nearly identical sequences at two or more points along the length of the Y chromosome. Together, these 285 multi-copy PCR assays identify a total of 706 MSY loci.
In total, the PCR assays for 1277 euchromatic STSs, 992 single-copy and 285 multi-copy, amplify 1698 loci distributed across the MSYs 23 Mb of euchromatic sequence. This corresponds to an average spacing of <14 kb between PCR-amplified loci (Table 1).
Most PCR assays cataloged in the database can be performed using one of two protocols:
(1) Standard PCR
94°C for 3 min
35 cycles of: 94°C for 1 min; 61°C for 1 min; 72°C for 1 min
72°C for 5 min
(2) Touchdown PCR
94°C for 1 min
20 cycles of: 92°C for 30 s; 70–0.5°C/cycle for 40 s (i.e. a ramp from 70 to 60.5°C with temperature decrements of 0.5°C per cycle)
20 cycles of: 92°C for 30 s; 60°C for 40 s + 1 s/cycle (i.e. a ramp from 40 to 59 s with time increments of 1 s per cycle)
Several PCR assays can also be performed using alternative protocols, as described in the associated NCBI GenBank entries.
QUERYING THE DATABASE
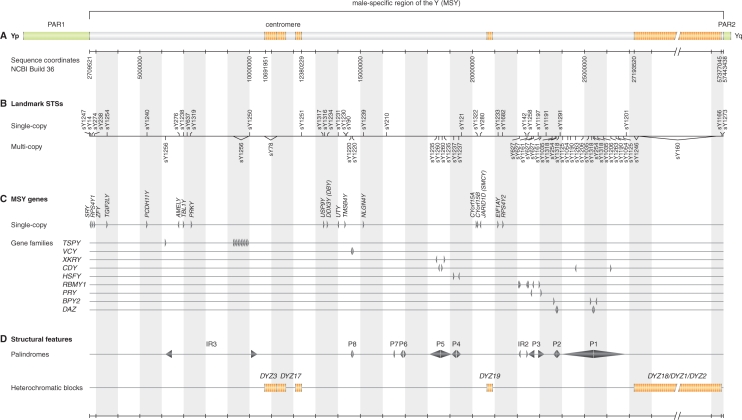
The query input webpage (http://breakpointmapper.wi.mit.edu/mapper.html) features a schematic of the human Y chromosome (Figure 1) that includes nucleotide sequence coordinates, landmark STSs, protein-coding genes and major structural features of the MSY. Sequence coordinates (Figure 1A) are based on NCBI Build 36, an assembly of the human genome sequence that incorporates all known human MSY sequence (1,38). The 51 landmark STSs (Figure 1B) consist of sY1247 and sY1273, located at the boundaries between MSY and pseudoautosomal sequences on Yp and Yq, respectively, and a previously published panel of 49 STSs that our laboratory has employed in screening for MSY deletions (see Table SM-2 in Supplementary Methods of Ref. 36). A table of these 51 landmark STSs is available on the query input webpage under Reference Tools (Figure 2A). MSY genes (Figure 1C) and structural features (Figure 1D) are annotated as previously described (1).
Figure 1.
The human MSY. (A) Schematic of the entire Y chromosome, with pseudoautosomal regions (PAR1 and PAR2) in green, euchromatic regions of MSY in gray and heterochromatic regions of MSY in orange. Immediately below are Y chromosome sequence coordinates based on NCBI Build 36. (B) Locations of 51 landmark STSs. (C) MSY protein-coding genes and gene families; arrowheads indicate 5′-to-3′ orientation. (D) Major structural features.
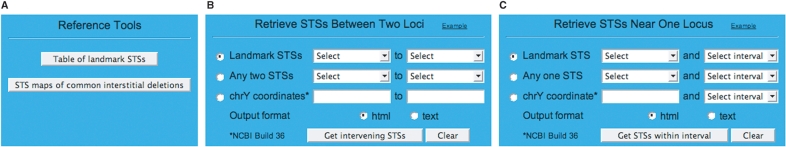
Figure 2.
Querying the database. (A) Reference Tools box containing links to a table of 51 landmark STSs and to deletion maps of common interstitial deletions. (B) Query box for generating a catalog of STSs between two landmark STSs, or between any two STSs in the database, or between any two Y chromosome sequence coordinates. (C) Query box for generating a catalog of STSs in the vicinity of a particular STS or sequence coordinate.
Two modes of querying the database are available to the user. The user may elect to generate a catalog of STSs between two loci (Figure 2B). They can be two landmark STSs, as listed in pull-down menus; any two STSs in the database, as listed in pull-down menus; or any two Y chromosome sequence coordinates, as entered by the user. Alternatively, the user may elect to generate a catalog of STSs in the vicinity of a particular STS or sequence coordinate (Figure 2C). In this case, the user selects the STS from a pull-down menu or enters the sequence coordinate, and then selects the size of the surrounding interval, again from a pull-down menu. Links to illustrations of these two modes of query are provided within the input boxes. In addition, a link to schematics of common MSY deletions, including predicted results for the 51 landmark STSs, is provided under Reference Tools (Figure 2A).
Regardless of the mode of query, the resulting catalog of STSs is displayed in two ways: as a table and in a custom Y chromosome browser (Figure 3). For each STS, the table indicates start and end sequence coordinates, PCR product length, primer sequences, amplification conditions and GenBank accession number. Multi-copy STSs are identified as such, and the sequence coordinates of each additional locus of amplification are tabulated. Below the table, a custom Y chromosome browser displays the region encompassing the cataloged STSs. The position of each STS and of each protein-coding gene within the interval is indicated. The user can also interrogate the Y chromosome browser independent of the initial STS database query.
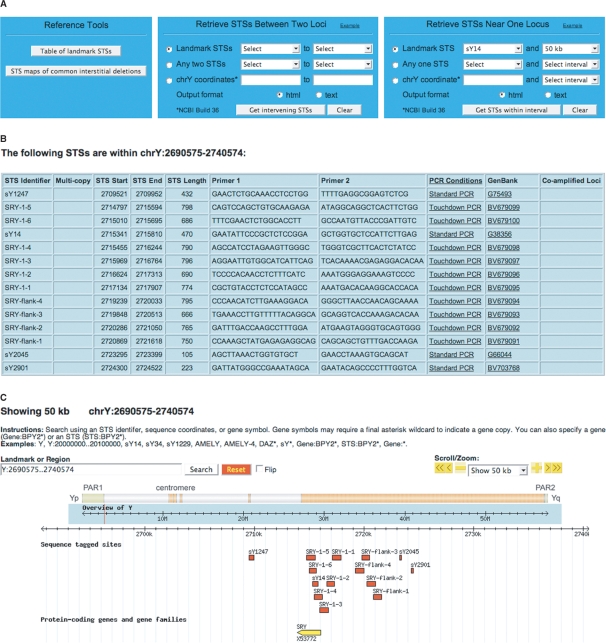
Figure 3.
An example of query results. (A) Querying the database for STSs in the 50-kb interval surrounding sY14. (B) The queried interval contains 14 STSs. For each STS, the table lists sequence coordinates, PCR product length, primers, PCR conditions (as a link to a pop-up), GenBank accession number (as a link to the NCBI entry), and in the case of a multi-copy STS, sequence coordinates of co-amplified loci. (C) The custom Y chromosome browser indicates the location of the queried interval, and the identities and positions of all STSs and protein-coding genes within the interval.
ACKNOWLEDGEMENTS
We thank Laura Brown, Simon Foote, Tomoko Kuroda-Kawaguchi, Bruce Lahn, Janet Marszalek, Tatyana Pyntikova, Renee Reijo Pera, Sjoerd Repping, Steve Rozen, Chao Sun, Charles Tilford and Douglas Vollrath for STS design and testing; and Jörg Gromoll for comments on the website. Supported by the National Institutes of Health and the Howard Hughes Medical Institute. Funding to pay the Open Access publication charges for this article was provided by the Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs PA, Ross A. Structural abnormalities of the Y chromosome in man. Nature. 1966;210:352–354. doi: 10.1038/210352a0. [DOI] [PubMed] [Google Scholar]
- 3.Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum. Genet. 1976;34:119–124. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- 4.Guellaen G, Casanova M, Bishop C, Geldwerth D, Andre G, Fellous M, Weissenbach J. Human XX males with Y single-copy DNA fragments. Nature. 1984;307:172–173. doi: 10.1038/307172a0. [DOI] [PubMed] [Google Scholar]
- 5.Page DC, de la Chapelle A, Weissenbach J. Chromosome Y-specific DNA in related human XX males. Nature. 1985;315:224–226. doi: 10.1038/315224a0. [DOI] [PubMed] [Google Scholar]
- 6.Vergnaud G, Page DC, Simmler MC, Brown L, Rouyer F, Noel B, Botstein D, de la Chapelle A, Weissenbach J. A deletion map of the human Y chromosome based on DNA hybridization. Am. J. Hum. Genet. 1986;38:109–124. [PMC free article] [PubMed] [Google Scholar]
- 7.Affara NA, Florentin L, Morrison N, Kwok K, Mitchell M, Cook A, Jamieson D, Glasgow L, Meredith L, et al. Regional assignment of Y-linked DNA probes by deletion mapping and their homology with X-chromosome and autosomal sequences. Nucleic Acids Res. 1986;14:5353–5373. doi: 10.1093/nar/14.13.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Affara NA, Ferguson-Smith MA, Tolmie J, Kwok K, Mitchell M, Jamieson D, Cooke A, Florentin L. Variable transfer of Y-specific sequences in XX males. Nucleic Acids Res. 1986;14:5375–5387. doi: 10.1093/nar/14.13.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson M, Page DC, de la Chapelle A. Chromosome Y-specific DNA is transferred to the short arm of X chromosome in human XX males. Science. 1986;233:786–788. doi: 10.1126/science.3738510. [DOI] [PubMed] [Google Scholar]
- 10.Disteche CM, Casanova M, Saal H, Friedman C, Sybert V, Graham J, Thuline H, Page DC, Fellous M. Small deletions of the short arm of the Y chromosome in 46, XY females. Proc. Natl Acad. Sci. USA. 1986;83:7841–7844. doi: 10.1073/pnas.83.20.7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petit C, de la Chapelle A, Levilliers J, Castillo S, Noel B, Weissenbach J. An abnormal terminal X-Y interchange accounts for most but not all cases of human XX maleness. Cell. 1987;49:595–602. doi: 10.1016/0092-8674(87)90535-6. [DOI] [PubMed] [Google Scholar]
- 12.Page DC, Brown LG, de la Chapelle A. Exchange of terminal portions of X- and Y-chromosomal short arms in human XX males. Nature. 1987;328:437–440. doi: 10.1038/328437a0. [DOI] [PubMed] [Google Scholar]
- 13.Levilliers J, Quack B, Weissenbach J, Petit C. Exchange of terminal portions of X- and Y-chromosomal short arms in human XY females. Proc. Natl Acad. Sci. USA. 1989;86:2296–2300. doi: 10.1073/pnas.86.7.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vollrath D, Foote S, Hilton A, Brown LG, Beer-Romero P, Bogan JS, Page DC. The human Y chromosome: a 43-interval map based on naturally occurring deletions. Science. 1992;258:52–59. doi: 10.1126/science.1439769. [DOI] [PubMed] [Google Scholar]
- 15.Weil D, Wang I, Dietrich A, Poustka A, Weissenbach J, Petit C. Highly homologous loci on the X and Y chromosomes are hot-spots for ectopic recombinations leading to XX maleness. Nat. Genet. 1994;7:414–419. doi: 10.1038/ng0794-414. [DOI] [PubMed] [Google Scholar]
- 16.Lahn BT, Ma N, Breg WR, Stratton R, Surti U, Page DC. Xq-Yq interchange resulting in supernormal X-linked gene expression in severely retarded males with 46,XYq-karyotype. Nat. Genet. 1994;8:243–250. doi: 10.1038/ng1194-243. [DOI] [PubMed] [Google Scholar]
- 17.Hsu LY. Phenotype/karyotype correlations of Y chromosome aneuploidy with emphasis on structural aberrations in postnatally diagnosed cases. Am. J. Med. Genet. 1994;53:108–140. doi: 10.1002/ajmg.1320530204. [DOI] [PubMed] [Google Scholar]
- 18.Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat. Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 19.Tsuchiya K, Reijo R, Page DC, Disteche CM. Gonadoblastoma: molecular definition of the susceptibility region on the Y chromosome. Am. J. Hum. Genet. 1995;57:1400–1407. [PMC free article] [PubMed] [Google Scholar]
- 20.Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Kohn FM, Schill WB, Farah S, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum. Mol. Genet. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- 21.Schiebel K, Winkelmann M, Mertz A, Xu X, Page DC, Weil D, Petit C, Rappold GA. Abnormal XY interchange between a novel isolated protein kinase gene, PRKY, and its homologue, PRKX, accounts for one third of all (Y+)XX males and (Y–)XY females. Hum. Mol. Genet. 1997;6:1985–1989. doi: 10.1093/hmg/6.11.1985. [DOI] [PubMed] [Google Scholar]
- 22.Sun C, Skaletsky H, Rozen S, Gromoll J, Nieschlag E, Oates R, Page DC. Deletion of azoospermia factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum. Mol. Genet. 2000;9:2291–2296. doi: 10.1093/oxfordjournals.hmg.a018920. [DOI] [PubMed] [Google Scholar]
- 23.Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum. Mol. Genet. 2000;9:2563–2572. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]
- 24.Blanco P, Shlumukova M, Sargent CA, Jobling MA, Affara N, Hurles ME. Divergent outcomes of intrachromosomal recombination on the human Y chromosome: male infertility and recurrent polymorphism. J. Med. Genet. 2000;37:752–758. doi: 10.1136/jmg.37.10.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat. Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 26.Repping S, Skaletsky H, Lange J, Silber S, Van Der Veen F, Oates RD, Page DC, Rozen S. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am. J. Hum. Genet. 2002;71:906–922. doi: 10.1086/342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Repping S, Skaletsky H, Brown L, van Daalen SK, Korver CM, Pyntikova T, Kuroda-Kawaguchi T, de Vries JW, Oates RD, et al. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat. Genet. 2003;35:247–251. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- 28.Fernandes S, Paracchini S, Meyer LH, Floridia G, Tyler-Smith C, Vogt PH. A large AZFc deletion removes DAZ3/DAZ4 and nearby genes from men in Y haplogroup N. Am. J. Hum. Genet. 2004;74:180–187. doi: 10.1086/381132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Repping S, van Daalen SK, Korver CM, Brown LG, Marszalek JD, Gianotten J, Oates RD, Silber S, van der Veen F, et al. A family of human Y chromosomes has dispersed throughout northern Eurasia despite a 1.8-Mb deletion in the azoospermia factor c region. Genomics. 2004;83:1046–1052. doi: 10.1016/j.ygeno.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 30.Jobling MA, Lo IC, Turner DJ, Bowden GR, Lee AC, Xue Y, Carvalho-Silva D, Hurles ME, Adams SM, et al. Structural variation on the short arm of the human Y chromosome: recurrent multigene deletions encompassing Amelogenin Y. Hum. Mol. Genet. 2007;16:307–316. doi: 10.1093/hmg/ddl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 32.Page DC. Hypothesis: a Y-chromosomal gene causes gonadoblastoma in dysgenetic gonads. Development (Cambridge, U.K.) 1987;101(Suppl.):151–155. doi: 10.1242/dev.101.Supplement.151. [DOI] [PubMed] [Google Scholar]
- 33.Nathanson KL, Kanetsky PA, Hawes R, Vaughn DJ, Letrero R, Tucker K, Friedlander M, Phillips KA, Hogg D, et al. The Y deletion gr/gr and susceptibility to testicular germ cell tumor. Am. J. Hum. Genet. 2005;77:1034–1043. doi: 10.1086/498455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foote S, Vollrath D, Hilton A, Page DC. The human Y chromosome: overlapping DNA clones spanning the euchromatic region. Science. 1992;258:60–66. doi: 10.1126/science.1359640. [DOI] [PubMed] [Google Scholar]
- 35.Tilford CA, Kuroda-Kawaguchi T, Skaletsky H, Rozen S, Brown LG, Rosenberg M, McPherson JD, Wylie K, Sekhon M, et al. A physical map of the human Y chromosome. Nature. 2001;409:943–945. doi: 10.1038/35057170. [DOI] [PubMed] [Google Scholar]
- 36.Repping S, van Daalen SK, Brown LG, Korver CM, Lange J, Marszalek JD, Pyntikova T, van der Veen F, Skaletsky H, et al. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat. Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 37.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 38.Kirsch S, Weiss B, Miner TL, Waterston RH, Clark RA, Eichler EE, Munch C, Schempp W, Rappold G. Interchromosomal segmental duplications of the pericentromeric region on the human Y chromosome. Genome Res. 2005;15:195–204. doi: 10.1101/gr.3302705. [DOI] [PMC free article] [PubMed] [Google Scholar]