Abstract
RegulonDB (http://regulondb.ccg.unam.mx/) is the primary reference database offering curated knowledge of the transcriptional regulatory network of Escherichia coli K12, currently the best-known electronically encoded database of the genetic regulatory network of any free-living organism. This paper summarizes the improvements, new biology and new features available in version 6.0. Curation of original literature is, from now on, up to date for every new release. All the objects are supported by their corresponding evidences, now classified as strong or weak. Transcription factors are classified by origin of their effectors and by gene ontology class. We have now computational predictions for σ54 and five different promoter types of the σ70 family, as well as their corresponding −10 and −35 boxes. In addition to those curated from the literature, we added about 300 experimentally mapped promoters coming from our own high-throughput mapping efforts. RegulonDB v.6.0 now expands beyond transcription initiation, including RNA regulatory elements, specifically riboswitches, attenuators and small RNAs, with their known associated targets. The data can be accessed through overviews of correlations about gene regulation. RegulonDB associated original literature, together with more than 4000 curation notes, can now be searched with the Textpresso text mining engine.
INTRODUCTION
A major current task for bioinformatics is that of representing biological information into an electronic and computable form. These computational representations make the large amounts of data amenable to analysis, integration and eventual transformation into knowledge. The integration and new way of understanding the biology of a single cell such as Escherichia coli K12 is a major challenge in genomics and should be a milestone for systems biology. RegulonDB is currently one of the largest database offering curated knowledge of the transcriptional regulatory network of any free-living organism (1). Escherichia coli K12 has about two-thirds of its total gene content experimentally characterized, whereas, for around a third, there is some information about their regulation. Our curation feeds both RegulonDB and EcoCyc (2) databases.
RegulonDB is constantly updated with information derived from original research papers. The second line in Table 1 provides the link to the summary table of the total curated objects throughout the years. Our own efforts on high-throughput experimental mapping of promoters, whose initial results are reported here, enabled a significant increase in the total number of experimentally identified transcription start sites (TSSs).
Table 1.
Links to RegulonDB and related resources
RegulonDB contains detailed information of the different elements that conform the known regulatory network of the cell, such as transcription factors (TFs), small RNAs (sRNAs) and operon structures with their associated regulatory elements: promoters, TF binding sites and terminators, and from this version on, attenuators, riboswitches and sRNA targets. The description of the network is also conceptually enriched by more precise definitions—for instance, simple and complex regulons (3) and regulator classes (global or local and internal or external sensing) as explained below. RegulonDB is complemented with computational analyses and genome-wide predictions of operons, promoters, TF binding sites, ribosome-binding sites and, for the first time, RNA regulatory target sites.
Visualizing tools in RegulonDB allow the user to navigate in the genome (Genome browser), to identify co-regulators for a particular TF, to locate the genes’ immediate neighbors in the regulatory network, and to identify sets of genes predicted to be functionally related (Nebulon tool). Moreover, it also incorporates tools for the analysis of the transcriptional regulation of global gene expression experiments made in E. coli K12 (GETtools), as well as for exhaustive analyses focused on the detection of regulatory signals in upstream regulatory regions (RSA tools).
This paper summarizes the modifications and improvements made during the last 2 years that are transforming RegulonDB into a more comprehensive computational model of regulation of gene expression in E. coli.
Enhanced and expanded description of regulatory elements of transcription initiation
RegulonDB is mainly a manual database of regulatory information in E. coli incorporated by a team of curators from the primary literature. PubMed abstracts are selected using a set of pertinent key words related to gene regulation. When there is direct or suspected new relevant information, the full text of the articles is analyzed and the data are added to RegulonDB and EcoCyc.
Starting on January 2008, every release of RegulonDB and EcoCyc will contain up-to-date curation with a delay of no more than 3 months. To achieve this, we have used three main curation strategies: by year, by regulon and sigmulon and by physiological system.
Classification of evidences (strong and weak)
The evidences associated to all RegulonDB objects are now classified as ‘strong’ or ‘weak,’ based on the confidence level of the experiment or prediction that supports objects and their relationships. A ‘strong’ evidence is assigned to an object when the experimental data provide high certainty of its existence; otherwise, it is a ‘weak’ evidence. Examples of strong evidences are DNA binding of purified TF for regulatory interactions, mapping of TSSs for promoters, and length of mRNA for transcription units. On the other hand, gene expression analyses and computational predictions are considered weak evidences. It is important to state that several weak evidences for an object do not become a strong one. These two types of evidences are distinguished graphically with solid or dashed lines for objects supported by strong or weak evidences, respectively.
Experimental TSS mapping
Experimental determination of TSSs provides primary information critical to identify promoters and regulatory regions that control gene expression. To expand beyond literature searches our knowledge of the regulatory universe of E. coli, we started a genome-wide project to experimentally map as many promoters as possible in this organism. For this purpose, we used a modified 5′RACE protocol (4) with gene-specific oligonucleotides. To validate the accuracy of this strategy, we determined the TSSs for 50 TUs, which have been previously published, 92% of these TUs showed a perfect match (with a discrepancy of up to one nucleotide with respect to the published TSS). The rest showed slight ambiguity inherent to the RACE protocol, of up to six nucleotides. We detected more than one TSS in 14 of these TUs. Interestingly, for only two of them, additional TSSs had been reported. Thus, our results are highly accurate and determine additional promoters to >25% of TUs previously determined.
A total of 317 TSSs for 269 TUs (38 have more than one TSS) had been mapped with the 5′RACE methodology. One hundred and ten of them correspond to TUs with hypothetical genes for which no function has been inferred. The newly mapped TSSs have been included in RegulonDB. A detailed compendium of these findings will be published elsewhere.
Computational prediction of promoters for alternative σ factors
We have generated computational predictions for four different promoters of the σ70 family: those of σ 24, 28, 32 and 38, in addition to the already existing σ70 promoters. Promoter predictions have also been generated for the σ54 factor, which defines a different σ factor family than σ70. The putative +1 of transcription initiation along with the −35 and −10 boxes can be downloaded from RegulonDB (see ‘Predicted promoters’ on Table 1). The method applied to generate the promoter predictions is described in (5) (see ‘Promoter analysis tools’ in Table 1).
Internal versus external sensing classes of transcription factors
The active and inactive conformation of TFs is regulated by specific cell signals (commonly called ‘effectors’) that can be metabolites, ions or other chemical-signaling molecules, through covalent or allosteric interactions. The origin of these effectors can be endogenous (synthesized inside the cell), exogenous (incorporated or transported from outside the cell) or both (hybrid). As proposed in (6), TFs are classified according to the origin of their effectors as internal, external, hybrid or unknown. This feature has been added to the TFs in the database and a link to a specific web page that shows details of the cell sensing properties of the transcriptional regulators has been created (Table 1).
All genes that code for known and predicted TFs have been annotated with their corresponding gene ontology class (7), and we uploaded those for the rest of the genes of the genome from EcoCyc.
RNA regulatory elements
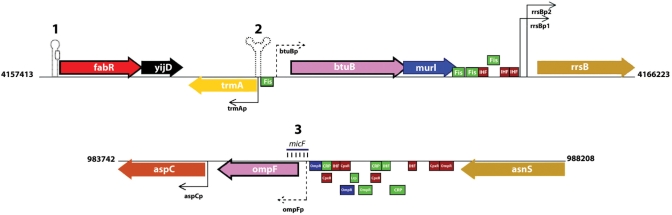
Until recently, regulation beyond transcription initiation was included in external tables. RegulonDB v.6.0 has an expanded conceptual and relational model that includes other levels and mechanisms of regulation of gene expression, such as transcriptional elongation, post-transcriptional modification and translational initiation. The first elements that are now modeled and populated are RNA regulatory elements, specifically riboswitches and attenuators, and small RNAs. The user interface has a graphic representation and textual information about their sequences, location, evidences and references; an example is shown in Figure 1 and tables containing all these data are available in the web pages indicated in Table 1.
Figure 1.
Graphic representation of the new objects in RegulonDB: 1. Attenuator, 2. Riboswitch, 3. sRNA.
Riboswitches and predicted attenuators
Riboswitches and attenuators are cis-regulatory elements that can modulate transcription elongation or translation initiation. A riboswitch is part of the 5′ non-translated region in specific bacterial mRNAs that can modulate gene expression in direct response to small molecules without the need for a protein intermediate. These regulatory elements are highly conserved, both in structure and sequence, probably due to the constraints of forming a highly structured binding pocket for the effector. Riboswitches are usually found associated with transcription or translation attenuators (8). Several of these riboswitches have been experimentally described and their sequences are obtained from Rfam, a database of RNA families (9). In addition to all known riboswitches, we have also added to RegulonDB all the other cis-regulatory RNA elements present in Rfam.
Attenuators are segments of RNA in the untranslated regions of some mRNAs that can form several mutually exclusive secondary structures, but, contrary to riboswitches, are rarely conserved at the sequence level. Under certain conditions, one of the structures will be the most stable, which will have a regulatory effect. Attenuators can act at the transcriptional level by causing the premature termination of transcription or at the initiation of translation by forming Shine-Dalgarno sequester structures (10–12). A set of more than 700 predicted attenuators (both transcriptional and translational) was generated by Merino et al. (13), taking into account the structural properties of known attenuators; these predictions are now included in RegulonDB.
Small RNAs
The sRNAs genes code for RNA sequences of <350 nucleotides long can have intrinsic catalytic activity (e.g. the 10S catalytic subunit of the RNase P), modify a protein activity (e.g. csrB/RNA, which binds to the CsrA translational regulator and thereby antagonizes its activity), or regulate the messenger stability or translational efficiency (e.g. micF, which binds to ompF mRNA to repress its translation) (14). RegulonDB now includes 49 interactions between sRNAs and their target genes.
Overviews of gene regulation biology and additional computational improvements
So far, we have provided two major mechanisms to access the knowledge available in RegulonDB: through the navigation of individual objects and their associated links and through the download of flat files with complete lists of objects and their properties (e.g. regulatory interactions, predicted promoters, terminators, operons, etc.). Now, we offer two new accession mechanisms: one through the download of the complete database (both data and schema can be downloaded in dump files to populate the most common database management systems such as MySQL, Postgress, Oracle and Apache Derby); and a new integrative level of description of the biology of gene regulation, with tables and graphs providing a collection of perspectives of different relationships and their distributions. For instance, these new tables and graphs help to identify how many and which genes are transcribed by each of the seven σ factors, the distribution of genes in operons, and the distribution of activator- and repressor-binding site positions (see ‘Data Overviews’ on Table 1).
RegulonDB v.6.0 has several computational and graphic user interface improvements: the graphic display of all the diagrams of genes and operons has been improved with a better quality and a better definition; the object names are completely visible inside the graph objects and mouse-over tooltips have been implemented on each object to simplify their identification by the user (e.g. binding site tooltips provide their central position); another improvement is the display of gene ontology. Automatic consistency checking for different objects has also been implemented to improve data integrity.
Implementation of the Textpresso text-mining engine
RegulonDB literature can now be searched with the Textpresso text-mining engine (15), customized for E. coli. Textpresso allows direct exploration of the curated literature, both at the level of highly specific key words and with entire categories or ontology classes (derived from GO concepts or customized word lists). The user can, for example, search for a type of regulation in which a gene or operon and a specific TF are mentioned within sentences of different papers. Currently, the tool can search through 2472 full-text papers, 3125 paper abstracts, and more than 4200 curator notes. The addition of this text-mining tool to RegulonDB will expand the possibilities, for the end user, of traversing the knowledge space of E. coli metabolism and gene regulation and will allow our curators to refine and confirm their annotations. See also (16).
New external links
In addition to the existing external database links (Swiss-Prot, GenBank, GenProtEC, OU MicroarrayDB), RegulonDB data is also accessible through EBI (17). We are also coordinated with the EcoliHub team to link RegulonDB as part of their wiki and integrated database tools (http://www.ecolihub.org).
Many functional insights can be gained by studying the context in which genes are conserved in different organisms. With this release of RegulonDB, we include a link to the Gene Context Tool (18) for every protein in the database, allowing the user to visualize the genomic context among all bacterial-sequenced genomes. As an example of the use of this tool, general function can, in many cases, be inferred for genes with no annotation by observing the neighboring genes of their orthologs in other bacteria.
DISCUSSION
One of the major challenges in bioinformatics is to provide concepts and methods that help to integrate large amounts of data in new and comprehensible ways. Our curation and modeling of gene regulation provide electronic and computable access to an important fraction of the vast amount of literature and knowledge for one of the best-studied organisms, the E. coli bacterium.
The relational model of RegulonDB has been expanded to include regulation beyond transcription and to incorporate concepts of gene regulation such as sigmulons and classes of TFs. In the future stimulons, modules and network motifs shall be included.
The value of this information is also enhanced by the links, formats, and availability to related resources, both related databases and useful programs for analysis and display. RegulonDB v.6.0 has new links to external resources (wiki EcoliHub, EBI) and new databases that access our curated knowledge (EcoGene, uniprot-genome summaries). Internally available resources have been expanded, particularly with the Textpresso-focused access to specific corpora of abstracts and papers from E. coli, links to TractorDB, and the dynamic overviews of correlations and distributions of regulation in E. coli.
ACKNOWLEDGEMENTS
We acknowledge Víctor del Moral for computer support, Alfredo Mendoza, Leticia Olvera and Maricela Olvera for promoter mapping. This work was supported by National Institutes of Health grant GM071962-03 and Universidad Nacional Autónoma de México grant 214905. Funding to pay the Open Access publication charges for this article was provided by the same grants.
Conflict of interest statement. None declared.
REFERENCES
- 1.Salgado H, Gama-Castro S, Peralta-Gil M, Diaz-Peredo, E, Sanchez-Solano F, Santos-Zavaleta A, Martinez-Flores I, Jimenez-Jacinto V, Bonavides-Martinez C, et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–D397. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutierrez-Rios RM, Rosenblueth DA, Loza JA, Huerta AM, Glasner JD, Blattner FR, Collado-Vides J. Regulatory network of Escherichia coli: consistency between literature knowledge and microarray profiles. Genome. Res. 2003;13:2435–2443. doi: 10.1101/gr.1387003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaefer BC. Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal. Biochem. 1995;227:255–273. doi: 10.1006/abio.1995.1279. [DOI] [PubMed] [Google Scholar]
- 5.Huerta AM, Collado-Vides J. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 2003;333:261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Antonio A, Janga SC, Salgado H, Collado-Vides J. Internal-sensing machinery directs the activity of the regulatory network in Escherichia coli. Trends Microbiol. 2006;14:22–27. doi: 10.1016/j.tim.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- 11.Dubnau D. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. CRC Crit. Rev. Biochem. 1984;16:103–132. doi: 10.3109/10409238409102300. [DOI] [PubMed] [Google Scholar]
- 12.Lovett PS. Translational attenuation as the regulator of inducible cat genes. J. Bacteriol. 1990;172:1–6. doi: 10.1128/jb.172.1.1-6.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merino E, Yanofsky C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 2005;21:260–264. doi: 10.1016/j.tig.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Storz G, Haas D. A guide to small RNAs in microorganisms. Curr. Opin. Microbiol. 2007;10:93–95. [Google Scholar]
- 15.Muller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez-Penagos C, Salgado H, Martinez-Flores I, Collado-Vides J. Automatic reconstruction of a bacterial regulatory network using Natural Language Processing. BMC Bioinformatics. 2007;8:293. doi: 10.1186/1471-2105-8-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kersey P, Bower L, Morris L, Horne A, Petryszak R, Kanz C, Kanapin A, Das U, Michoud K, et al. Integr8 and Genome reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciria R, Abreu-Goodger C, Morett E, Merino E. GeConT: gene context analysis. Bioinformatics. 2004;20:2307–2308. doi: 10.1093/bioinformatics/bth216. [DOI] [PubMed] [Google Scholar]