Abstract
Norine is the first database entirely dedicated to nonribosomal peptides (NRPs). In bacteria and fungi, in addition to the traditional ribosomal proteic biosynthesis, an alternative ribosome-independent pathway called NRP synthesis allows peptide production. It is performed by huge protein complexes called nonribosomal peptide synthetases (NRPSs). The molecules synthesized by NRPS contain a high proportion of nonproteogenic amino acids. The primary structure of these peptides is not always linear but often more complex and may contain cycles and branchings. In recent years, NRPs attracted a lot of attention because of their biological activities and pharmacological properties (antibiotic, immunosuppressor, antitumor, etc.). However, few computational resources and tools dedicated to those peptides have been available so far. Norine is focused on NRPs and contains more than 700 entries. The database is freely accessible at http://bioinfo.lifl.fr/norine/. It provides a complete computational tool for systematic study of NRPs in numerous species, and as such, should permit to obtain a better knowledge of these metabolic products and underlying biological mechanisms, and ultimately to contribute to the redesigning of natural products in order to obtain new bioactive compounds for drug discovery.
INTRODUCTION
Nonribosomal peptides (NRPs) show various particularities and a large structural diversity. They are short (2 to about 50 amino acids) and contain nonproteogenic amino acids. Indeed, amino acids other than the classical 20 ones found in proteins can be incorporated into those peptides. In addition, their particular way of synthesis can lead to chemical modifications of incorporated residues such as epimerization or methylation. Products from other biosynthesis pathways such as lipids or carbohydrates can also be introduced. The NRPs thus show a great diversity of their monomer composition. The primary structure of the NRPs is not always linear but often more complex: they can be linear like classical ribosomal peptides, but also branched, cyclic (partially or totally) or even poly-cyclic. Structural and compositional variety of these peptides allows them to have a broad range of important biological and pharmacological activities. For example, the ACV-tripeptide, the famous penicillin and cephalosporin precursor, is synthesized by this way. NRPs show immunosuppressive (cyclosporine), antitumor (bleomycin) or antibiotic (vancomycine) activities. Other examples include siderophores (pyoverdin), toxins (HC-toxin) or surfactants (surfactin).
NRPs are synthesized by large enzymatic complexes called nonribosomal peptide synthetases (NRPSs). This mechanism has been described for the first time in 1971, during the study of two antibiotics: gramicidin S and tyrocidin (1). A NRPS represents at the same time a template and biosynthetic machinery (2). Genes coding for NRPS are organized in operons or in clusters. NRPSs are modularly organized. Each module is responsible for the incorporation of a specific monomer. Modules are subdivided into domains, each domain catalyzing a specific reaction in the incorporation of a monomer. Four main domains are necessary for a complete synthesis. The first one, the adenylation domain, selects and activates the monomer transforming it into adenylate form. The thiolation or peptidyl carrier protein domain covalently binds the activated monomer to the synthetase. The condensation domain catalyses the peptide bond formation between the residues linked onto two adjacent modules. Finally, the thioesterase domain, only present in the final module, releases the peptide from the synthetase. The product can either be released as a linear compound or get transformed into a cyclic peptide through an intramolecular reaction. In NRPS of iterative type, the thioesterase domain can allow the enzyme to iterate the collinear biosynthesis several times.
Secondary domains that allow residue modifications are present in many NRPSs. For example, an epimerization domain leading to obtaining the D isomer of an amino acid can be encountered. Methylation, oxidation or cyclization domains can also be found in some NRPSs.
The NRPS mechanism can produce different variants of a peptide that have the same structure but have different monomers at certain positions. It has been shown that variations of fermentation broths can lead to production of more than 30 cyclosporine variants (3). In other cases, variant synthesis is due to a diversity of genomic sequences. For example, it has been shown that the DNA sequences of NRPSs that produce the bacillomycin D (4) and bacillomycin L (5) variants are different. A better knowledge of the biosynthetic mechanism opens a way to redesign natural products and to obtain new bioactive peptides for drug discovery (6).
When the NRPS mechanism has been discovered, it seemed to be of little significance. It appeared to be more and more important in the literature due to the discovery of numerous genes coding for NRPSs and important biological activities of their products. Currently, there are still few research groups that develop methods or computational tools for manipulating NRPs. Among existing resources, the NRPS-PKS database (7) is focused on the synthetases and contains only 20 or so peptides. Other resources like PubChem (8) contain some NRPS peptides as well as other small biological molecules but only few variants are presented. The Peptaibol Database (9) is focused only on a specific family of non-ribosomal products. A comprehensive resource compiling all known NRPs has been missing so far.
To fill this lack, we developed the Norine database containing a large amount of NRPS peptides with all types of structure and activity. The name Norine stands for NOnRibosomal peptides, with ine as a typical ending of NRP names. The database currently contains more than 700 peptides and this number is still growing. Norine is freely available at http://bioinfo.lifl.fr/norine/.
CONTENTS
Several reviews describing the NRPS mechanism have been published (2,10–13). These publications contain some examples of peptides produced by this way but no resource including an exhaustive up-to-date list of NRPs has been available so far. We explored relevant papers published since 1970s to compile an exhaustive list of known NRPs. The Norine database currently features more than 700 peptides extracted from about 350 publications. All data of Norine comes from the scientific literature and has been manually curated (predicted data are not included), which insures the reliability of the annotations. Various types of annotations are stored in the database.
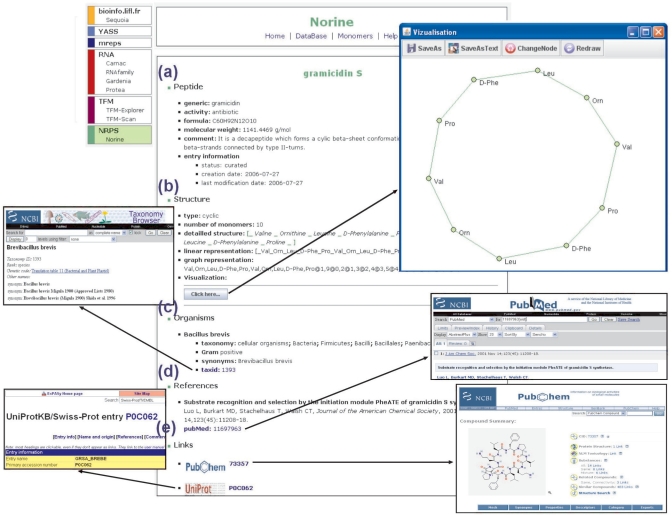
Figure 1 provides a representative screenshot showing the description of a peptide. The web page is organized into several parts. The first part, entitled ‘peptide’ (Figure 1a), presents general annotations of the peptide such as the peptide name and its synonyms. This is followed by fields presenting known biological activities of the peptide molecule, its molecular formula with the associated molecular weight and a possible comment presenting additional information on the molecule. Finally, the ‘entry information’ field contains the peptide status (curated or putative nonribosomal product), and creation and last modification dates that allow the user to follow the history of the entry.
Figure 1.
Description page of a peptide. The page is obtained when a peptide is selected after a search in the database. (a) Part ‘Peptide’ contains general information on the peptide. (b) Part ‘Structure’ compiles all the information on the peptide structure. A Java applet allows the peptide visualization in two-dimensions. (c) Part ‘Organisms’ concerns the organisms known to produce the peptide. Via a link, the user can access to NCBI taxonomy. (d) Part ‘References’ contains bibliographical references associated to the peptide. The user can access the corresponding entry of NCBI PubMed through a link. (e) Part ‘Links’ contains links to NCBI PubChem and UniProt.
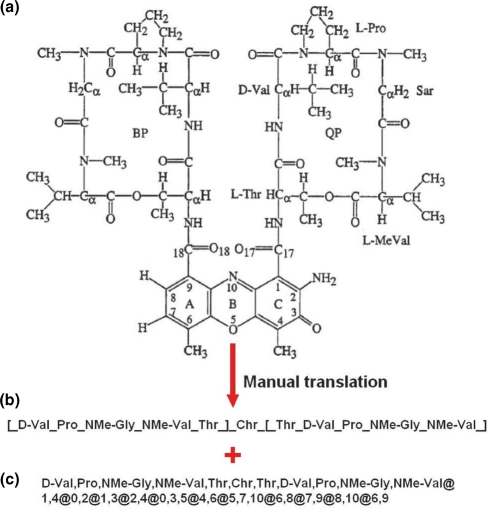
The next ‘structure’ section (Figure 1b) contains the most original data stored in Norine: structural features of the peptide. We chose to represent the peptide structure at the monomeric level rather than use a classical chemical atomic representation. This choice is justified by the fact that NRPs are synthesized by successive addition of monomers and not by atomic reactions, and therefore representing a peptide by its monomeric structure is an adequate way of specification. The first information found in this part is the peptide structural type. In Norine, the NRPs are classified in several groups according to their structural type: linear, branched, cyclic, partial cyclic, double cyclic and other. The group ‘other’ contains peptides that show a complex structure with several overlapping cycles and branches. The number of monomers composing the peptide is also given. The peptide structure is then presented using two representations. The first one is the ‘linear representation’ (Figure 2b). We developed this representation as a quick and easy way to represent a (possibly nonlinear) monomeric structure of the peptide by a linear string. In this representation, monomers are encoded according to a set of simple rules. The 20 proteogenic amino acids are encoded by the classical three-letter code (for example, Ala for alanine). When a functional group (like methyl) is added, its symbol is also added of the three-letter code (for example, NMe-Ala for N-methyl-alanine). By default, the amino acid is in l-form, the d-symbol is added when it is in d-form. To represent the structure, chained monomers are separated by an underscore sign. Cycles and branchings are represented, respectively, by brackets and braces. Note that this representation does not specify whether the bound involves only the backbone atoms of the monomer or side chain atoms. An example of linear representation of a double cyclic structure is given in Figure 2b. The linear representation provides a fast way of specifying a large class of structures using a set of simple rules. This class contains structures with no overlapping cycles covering a broad range of practical cases. However, structures with overlapping cycles cannot be specified unambiguously by a linear representation.
Figure 2.
Representation of NRP structures in Norine. (a) Chemical representation of actinomycin D (16). (b) Linear representation of actinomycin D. Monomers are encoded and separated by an underscore. Cycles are represented by brackets. (c) Graph representation of actinomycin D. It starts with a list of monomers each of which is associated with its rank in the list (numbered from zero) and corresponds to a node of the graph. Then, the adjacency list represents the edges incident to each node.
Another representation is called the ‘graph representation’ (Figure 2c). In this case, a peptide is represented as an undirected graph with nodes labeled by monomers and edges corresponding to the bonds between monomers. However, a standard computer representation of graphs (such as adjacency lists) does not allow the user to quickly figure out its 2D image. We thus developed a Java applet that draws the peptide structure in two dimensions. This applet is based on the Fruchterman–Reingold graph layout algorithm (14) that avoids edge crossing and keeps uniform edge lengths. The users can save the peptide structure in an image or text format, redraw the structure or still switch the node representation.
The annotations in the ‘organisms’ section (Figure 1c) concern the producing organisms. In general, the same peptide can be produced by several organisms and one organism can produce several NRPS peptides. Different data about these organisms are included: the organism name and its synonyms, taxonomic information, the Gram type for bacteria, a link to the NCBI taxonomy via the NCBI taxonomy identifier (taxid) (8).
The ‘references’ section (Figure 1d) includes bibliographical references associated to the peptide. For each reference, its title, authors, year of publication, journal name and NCBI PubMed link (8) are given.
The last part, the ‘links’ section (Figure 1e), includes links to other databases. A link to UniProt (15) allows the user to obtain information on synthetase such as its amino acid sequence. A link to NCBI PubChem compound (8) gives a direct access to the peptide chemical information.
WEB INTERFACE
The data are stored in a relational database using PostgreSQL database system. It can be queried via a friendly web interface in order to select NRPs corresponding to various search criteria. The web interface has been implemented with Java Server Page (JSP) technology using the Apache tomcat server (http://tomcat.apache.org/).
Two types of search are available: a general search and a structure search.
General search
General search can be done in different ways. The first one, called ‘basic search’, allows the user to search peptides according to several fields. The user can search by general or specific peptide names. A specific name represents the exact name of an NRP (e.g. cyclosporine A). If the peptide is present in the database, only one result will be obtained in this case. A general name represents a generic name for NRP (e.g. cyclosporine). This kind of search can result in several peptide variants belonging to the same group. The two names can be combined by any one of the Boolean operators AND, OR, AND NOT. Other search criteria include the biological activity and the structural type. When several fields are selected (such as name and activity) the results must match each of them. Peptides can also be searched by their molecular weight. To do this, the user has to specify an interval of possible molecular weights. The whole list of peptides is accessible by clicking a button.
‘Reference search’ allows a search for peptides by their bibliographical references. One can search by author, title, year of publication, journal or by pmid (NCBI PubMed identifier). Once again, it is possible to combine two reference fields with a Boolean operator.
In ‘organism search’, the search is done by organisms known to produce the corresponding peptides. The user can specify a desired taxonomic level. For example, querying ‘bacteria’ will output all the peptides produced by bacteria, and querying ‘Bacillus subtilis’ will output only those produced by this organism. Boolean operators can be used to combine two search fields. The complete list of organisms included in the database can be obtained by clicking a button.
Structure search
As structural variability is an important characteristic of NRPs, particular attention was given to ‘structure search’ that allows the user to search for peptides that verify certain structural properties. Two types of structure search have been implemented.
The first one is ‘composition-based search’, which looks up for peptides according to monomer composition features. As a simplest example, one can obtain all the peptides which contain a specific monomer, by specifying the corresponding monomer identifier. For example, querying Thr (for threonine) yields the list of all the peptides that contain at least one threonine. One can also obtain all the peptides containing a given monomer and its derivates. For example, querying Thr in this case gives all the peptides containing threonine amino acid, but also those that contain allo-threonine, d-threonine or choro-threonine. The user can search for peptides containing a given number of monomers, or less than or greater than a given number. Finally, the user can search for peptides containing a given list of monomers, either all of them or with a given maximum number of ‘errors’. This search does not take into account the peptide structure but only its monomeric composition. For example, querying <Ala,Pro,Val,Gly,Pro> with two possible errors returns all the peptides containing all the five monomers but also the peptides that contain only four (one error) or three (two errors) of the five monomers given in entry. Note that the query list can contain the same monomer several times.
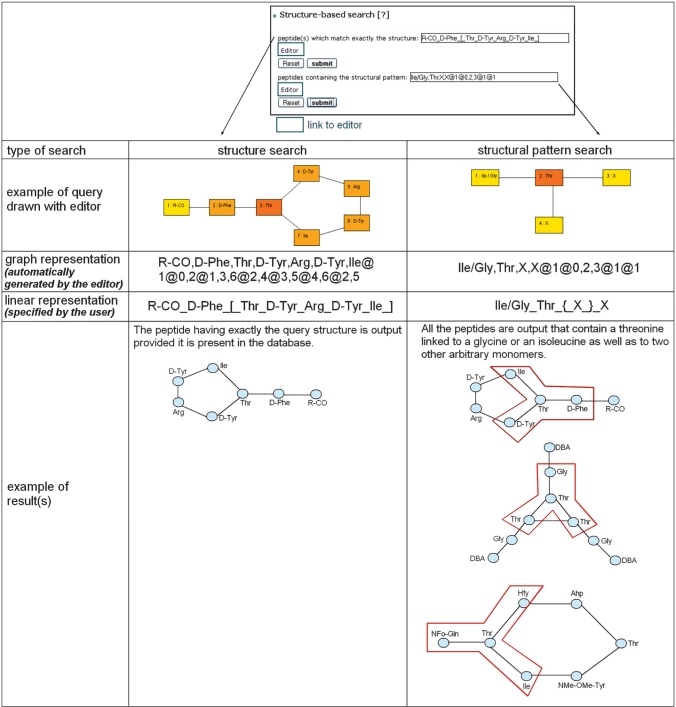
The user can also search for a peptide by specifying its structure in the ‘structure-based search’ (Figure 3). The peptide structure can be specified using either a linear or a graph representation. A graph representation can be created using a dedicated structure editor integrated to Norine. The Norine editor is a Java applet that allows one to build quickly and easily a graph representation of a peptide, i.e. to specify monomers and links between them. Complex monomeric structures can be easily drawn with a friendly graphical interface: first, monomers are selected and corresponding graph nodes are created, which are then connected by drawing edges between the nodes. It is also possible to delete some monomers or the whole structure. The user can also open a text file generated by the visualization applet in order to modify the created graph by hand. Once the structure is completed, clicking the ‘go’ button returns the peptide structure to the appropriate field of the Norine search page and the search for it in the database can be launched right away. Both with the linear and graph representations, the user can specify either the entire peptide structure to look for, or a structural pattern that the peptide must contain. The containment is defined as the usual subgraph relation: a pattern occurs in a peptide if each node of the pattern labeled by a monomer can be associated to a node of the peptide labeled by the same monomer so that the linked (unlinked) nodes of the pattern are linked (respectively unlinked) in the peptide.
Figure 3.
Example of structure-based search. Two search features are provided. The structure search looks up for a peptide having exactly the query structure. The structural pattern search looks up for the peptides containing the query pattern as a subgraph. The query pattern can contain joker or alternatively-labeled nodes (X and/). In both types of search, the query can be specified using either linear or graph representation. A link to the dedicated peptide structure editor (in green) allows the user to automatically obtain the graph representation. Alternatively, the user can specify the query through the linear representation. In the last row, examples of resulting peptides are given.
A structural pattern is specified in the same way as a regular peptide, i.e. using a linear or graph representation, where the latter is specified with the Norine structure editor. However, these representations are enriched by the possibility for a structural pattern to contain nodes labeled by several alternative monomers. These can be specified by a list of monomers separated by a special ‘/’ symbol. A special ‘X’ symbol stands for any monomer. The occurrence of patterns having alternatively labeled nodes is defined in the natural way.
CONCLUSION AND PERSPECTIVES
Nonribosomal synthesis is an original biosynthesis pathway that leads to a great diversity of products. A huge structural diversity of the NRPs allows them to have a broad range of important biological activities. This work resulted in compiling the first database entirely dedicated to the peptides produced by NRPS. Norine already contains more than 700 peptides and will continue to be completed and regularly updated.
Different features of Norine, and in particular different types of queries the user can make to the database, lead to different possibilities of its usage. In general, the user can easily extract different types of information about known NRPs. For example, Norine can be used to identify a peptide predicted by some other means from an NRPS amino acid sequence. One can then determine if the predicted peptide has already been identified by using the structure search features of Norine. Various other types of information can be extracted from the Norine database.
We expect that structure comparison tools can help to better understand the structure/function relationships of NRPs. More generally, the possibilities of study of different properties of peptides offered by Norine can bring new insights on their impact to their biological activity. We also believe that possibilities provided by Norine, in association with other NRPS enzyme dedicated tools, can lead to facilitate the redesign of natural products in order to develop new bioactive compounds for drug discovery. Indeed, combinatorial biosynthesis, the process of genetic manipulations of natural product biosynthetic machinery, depends, in particular, on the detailed knowledge of the involved metabolic processes. The product data contained in Norine should permit to better identify the specificity of different domains and to facilitate the search for domains incorporating a given residue. Note that few NRPS sequences are currently available in comparison with the number of peptide products.
In near future, we plan to enrich Norine with new computational tools such as the search for similarities between a new or unknown peptide and those already present in the database.
ACKNOWLEDGMENTS
This work was supported by PPF bioinformatique of Lille. S.C. was supported by an INRIA/Région Nord-Pas-de-Calais fellowship. ProBioGEM lab is supported by the region Nord-Pas-de-Calais, the Ministere de l’Enseignement et de la Recherche (ANR) and the European Funds for the Regional Development. Funding to pay the Open Access publication charges for this article was provided by INRIA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lipmann F, Gevers W, Kleinkauf H, Roskoski R. Polypeptide synthesis on protein templates: the enzymatic synthesis of gramicidin S and tyrocidine. Adv. Enzymol. Relat. Areas Mol. Biol. 1971;35:1–34. doi: 10.1002/9780470122808.ch1. [DOI] [PubMed] [Google Scholar]
- 2.Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem. Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 3.von Döhren H. Biochemistry and general genetics of nonribosomal peptide synthetases in fungi. Adv. Biochem. Eng. Biotechnol. 2004;88:217–264. doi: 10.1007/b99262. [DOI] [PubMed] [Google Scholar]
- 4.Moyne AL, Cleveland TE, Tuzun S. Molecular characterization and analysis of the operon encoding the antifungal lipopeptide bacillomycin D. FEMS Microbiol. Lett. 2004;234:43–49. doi: 10.1016/j.femsle.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Hofemeister J, Conrad B, Adler B, Hofemeister B, Feesche J, Kucheryava N, Steinborn G, Franke P, Grammel N, et al. Genetic analysis of the biosynthesis of non-ribosomal peptide and polyketide-like antibiotics, iron uptake and biofilm formation by Bacillus subtilis. A1/3. Mol. Genet. Genomics. 2004;272:363–378. doi: 10.1007/s00438-004-1056-y. [DOI] [PubMed] [Google Scholar]
- 6.Van Lanen SG, Shen B. Progress in combinatorial biosynthesis for drug discovery. Drug Discov. Today. 2006;3:285–292. doi: 10.1016/j.ddtec.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Ansari MZ, Yadav G, Gokhale RS, Mohanty D. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 2004;32(Web Server issue):W405–W413. doi: 10.1093/nar/gkh359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35(Database issue):D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitmore L, Wallace BA. The peptaibol database: a database for sequences and structures of naturally occurring peptaibols. Nucleic Acids Res. 2004;32(Database issue):D593–D594. doi: 10.1093/nar/gkh077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grünewald J, Marahiel MA. Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides. Microbiol. Mol. Biol. Rev. 2006;70:121–146. doi: 10.1128/MMBR.70.1.121-146.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Challis GL, Naismith JH. Structural aspects of non-ribosomal peptide biosynthesis. Curr. Opin. Struct. Biol. 2004;14:748–756. doi: 10.1016/j.sbi.2004.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller U, Schauwecker F. Combinatorial biosynthesis of non-ribosomal peptides. Comb. Chem. High Throughput Screen. 2003;6:527–540. doi: 10.2174/138620703106298707. [DOI] [PubMed] [Google Scholar]
- 13.Schwarzer D, Finking R, Marahiel MA. Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 2003;20:275–287. doi: 10.1039/b111145k. [DOI] [PubMed] [Google Scholar]
- 14.Fruchterman TMJ, Reingold EM. Graph drawing by force-directed placement. Software Pract Exper. 1991;21:1129–1164. [Google Scholar]
- 15.niProt Consortium1. The universal protein resource (UniProt) Nucleic Acids Res. 2007;35(Database issue):D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Liu X, Patel DJ. DNA bending and unwinding associated with actinomycin D antibiotics bound to partially overlapping sites on DNA. J. Mol. Biol. 1996;258:457–479. doi: 10.1006/jmbi.1996.0262. [DOI] [PubMed] [Google Scholar]