Abstract
Gene3D provides comprehensive structural and functional annotation of most available protein sequences, including the UniProt, RefSeq and Integr8 resources. The main structural annotation is generated through scanning these sequences against the CATH structural domain database profile-HMM library. CATH is a database of manually derived PDB-based structural domains, placed within a hierarchy reflecting topology, homology and conservation and is able to infer more ancient and divergent homology relationships than sequence-based approaches. This data is supplemented with Pfam-A, other non-domain structural predictions (i.e. coiled coils) and experimental data from UniProt. In order to enhance the investigations possible with this data, we have also incorporated a variety of protein annotation resources, including protein–protein interaction data, GO functional assignments, KEGG pathways, FUNCAT functional descriptions and links to microarray expression data. All of this data can be accessed through a newly re-designed website that has a focus on flexibility and clarity, with searches that can be restricted to a single genome or across the entire sequence database. Currently Gene3D contains over 3.5 million domain assignments for nearly 5 million proteins including 527 completed genomes. This is available at: http://gene3d.biochem.ucl.ac.uk/
INTRODUCTION
The identification of structural domains and their homologous relationships, and hence the domain composition of protein sequences, allows both the practical application of powerful approaches for functional prediction and theoretical investigations into protein structure evolution. Several resources exist to support this field, each bringing a particular perspective: the most comprehensive is InterPro (1)—an amalgamation of resources that links well to the UniProt (2) sequence database. Pfam (3) is the single largest resource and is of interest since it primarily classifies domains through the creation of sequence families. This means that it can be of particular use in functional association studies, though it does miss many ancient evolutionary relationships. Also, and in some ways most similar to Gene3D, there is the Superfamily database (4), which provides the SCOP-derived domain assignments (5) for genomic sequences.
Gene3D is designed to extend the CATH (6) structural domain database from the wwPDB (7) to the protein sequence databases UniProt and RefSeq (8) and the completed genomes defined by Integr8 (9). CATH domains are manually classified following automated analyses of the PDB and assigned a place in the CATH structural hierarchy, reflecting their structural composition and evolutionary relationships. To predict sequence relatives for these CATH domains, sets of hidden Markov Models (HMMs) are generated to represent each CATH superfamily. By scanning these HMM models against the sequence resources and resolving the ‘hits’, Gene3D v6.0 provides >3.5 million CATH domain assignments for nearly 2.5 million distinct proteins, including 49% of UniProt and 47% of complete genomes. The protocol for doing this has been previously described in (10) and is also illustrated in Supplementary Figure 1.
To further enhance these investigations Gene3D also integrates other domain predictions (i.e. Pfam-A), PDB-based assignments directly from CATH, several function resources [i.e. GO (11)] and protein–protein interaction (PPI) data [i.e. IntAct (12)]. All protein sequences are also clustered into hierarchical protein families to facilitate functional grouping of sequences.
By creating a simple interface between these resources it is now possible to examine in detail functional and evolutionary changes in relation to structures and to enhance functional and structural annotation of genomes. Gene3D has been a significant aid in selecting targets for the Structural Genomics Initiative (13). Over the last two years several new resources have been added and new methods of accessing the data made available. Foremost are the significant improvements made to the usability and functionality of the website, expansion of the pre-made sets available for download and the implementation of a suite of DAS servers using ProServer (14). These developments and their application are described in detail below.
GENE3D V6.0
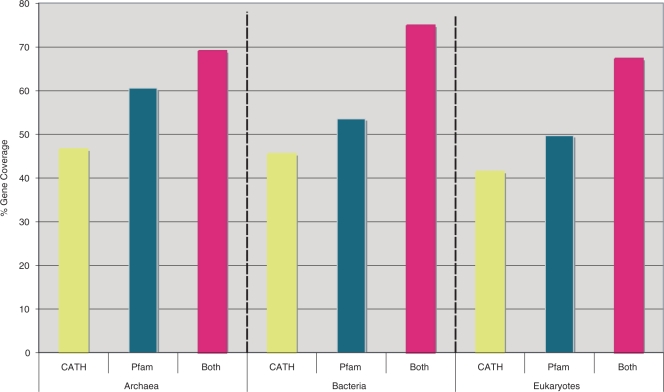
The September 2007 version of Gene3D contains ∼4.5 million distinct proteins, grouped into 190 000 protein families with more than 5 members (method described below)—around 600 000 proteins remain as ‘singletons’. Included in this are also 527 species (676 strains)—50 eukaryotes, 437 eubacteria and 39 archaea—totalling ∼1.9 million distinct proteins. See Figure 1 for the coverage of these genomes with CATH and Pfam domains. All the HMM-identified domains assigned to the 2046 CATH v3.1.0 superfamilies are sub-clustered at ten discrete sequence identity levels, ranging from 30–95% (files available for download), so as to aid accurate function transfer. For further details on additional annotation, including Pfam, low complexity regions, coiled coils, transmembrane helices, see Supplementary Table 2.
Figure 1.
Gene coverage of completed genomes in Gene3D. Shown in this figure are the percentages of genes in bacteria, archaea and eukaryotes that have at least one domain assigned by either (A) CATH, (B) Pfam or (C) both. It should be noted that not all the genomes have been completely scanned with Pfam—hence the coverage is lower than would be expected.
Functional data is represented through the inclusion of the GO, KEGG (15), COGs (16) and FunCat (17) datasets. Gene3D also has protein–protein interaction (PPI) data sourced from IntAct, MINT (18) and manually curated high-quality interactions from MPact (19) PPI datasets and where possible proteins are linked to expression data at ArrayExpress (20). For a complete list of imported resources, see Supplementary Table 3. We also aim to import and enable the use of as many different types of identifier as possible: currently the website can be queried with more than 35 million distinct identifiers sourced from UniProt, CATH, Pfam, the wwPDB, RefSeq, COGs, OMIM (21), BioThesaurus (22) and more (see Supplementary Table 1).
Changes to structural data
We have incorporated the manually curated PDB-based CATH v3.1.0 domain assignments (88 774 out of 93 885) by mapping them to UniProt using the procedure described by Andrew Martin (23). Multi-Domain Architectures (MDAs) were fully resolved for 7591 proteins out of 8646 possible. The resolution is carried out very conservatively and if any mapping problems between PDB and UniProt are identified the MDA is not calculated. Hence, this set can be considered a gold standard for structural annotation of UniProt.
CATH have also added ‘unassigned’ domains to their structural library. These are domains that have been identified within newly determined multidomain structures, but not yet classified in the CATH hierarchy. We also scan HMMs based on these to extend the possible structural coverage. In Gene3D v6.0, these add ∼250 000 domain assignments to ∼160 000 proteins.
The UniProt protein files are also a rich source of experimentally determined structural information and we now directly import various features including: signal peptides, active sites, metal-binding sites, splice sites and disulphide bonds. A collaboration with the BIOSAPIENS/ENCODE consortia exploiting this data revealed that 5–20% of human genes produced transcripts that exhibit some form of domain insertion, deletion or substitution whilst still remaining potentially functional (24).
Changes to interaction data
Protein–protein interaction (PPI) data is now sourced from the comprehensive MINT and IntAct resources, as well as the yeast-specific manually curated subset of MPact.
New whole chain families
One of the primary focuses of our research is to extend experimentally derived molecular studies to the vast number of experimentally uncharacterized proteins through bioinformatic methods. One of the most powerful means of carrying this out is through reliable functional inheritance between similar proteins. Various studies have shown that knowledge of domain architecture and sequence similarity can enable reliable transference of functional annotation (25,26). To enhance these approaches every protein in the database is assigned to a family based on sequence similarity. A novel approach has been employed that takes advantage of the fast affinity propagation clustering (APC) algorithm (27) and the comprehensive protein sequence similarity database SIMAP (28).
The Gene3D clustering protocol consists of several steps that aim to break down the problem of clustering 4.6 million sequences. The sequence database is clustered repeatedly, currently with fairly conservative thresholds (E-value 0.001, overlap length 80%), using the cd-hit (29) program and a mixture of single-linkage, multi-linkage and APC clustering. Ultimately, each derived cluster is subclustered at 10 levels of sequence identity. This quicker process should allow for improved benchmarking and analysis. For full details of the process, see Supplementary Data.
New identifier mappings
As mentioned above, we import and map identifiers from many new resources. This allows improved querying of and linking to Gene3D, as well as correlating disparate datasets. The new identifiers include SGD (Saccharomyces Genome Database) and most of those in the BioThesaurus database (i.e. OMIM and Ensembl identifiers). If an identifier maps to multiple proteins—either because several genes have been given the same name or because the identifier corresponds to a family—then all proteins are returned allowing the user to choose the one(s) of interest. However, as described below there are ways of refining the query or specifying particular (i.e. functional) subsets.
Hierarchical phylogenetic domain profiles—PhyloTuner
As mentioned above, all CATH superfamilies are subclustered at ten levels of sequence similarity. From these clusters phylogenetic profiles at each level of similarity are generated for the complete genomes from Integr8. By using the actual copy number of occurrences of each domain at each identity level it is possible to identify co-evolving domain families or subfamilies, as exemplified by the PhyloTuner approach developed by Ranea et al. (30). These profiles are provided on the FTP site, linked to at the top of the website pages. The advantage of this type of approach for functional prediction is that it can detect entirely novel associations that have no previous experimental evidence and hence guide the discovery of new knowledge rather than extending what is already known.
SIMPLER, MORE POWERFUL WEBSITE
Improved interface
Considerable effort has been put into improving the usability of the website and query results are now presented more swiftly in a clearer format. The primary focus of the site is on searching the database with an identifier term (i.e. CATH superfamily code) and returning the proteins associated with that term (i.e. members of that superfamily). The results are returned as a single page containing a set of selectable tabs for the functional, structural and taxonomic information associated with the query. The main view is common to every query, while the tab content can vary depending on whether the query returns a single protein or multiple proteins. Within the tabs, individual identifiers that can be searched in Gene3D are marked with a twisting arrow tag; clicking on this tag will submit the query.
Sophisticated aided querying
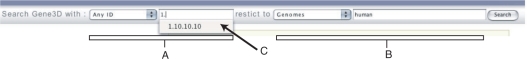
One of the key enhancements has been the construction of a much more sophisticated query tool (Figure 2). It takes the form of a bar across the top of the page containing a series of boxes for entering different options. The first two boxes are for specifying the identifier (i.e. the query term); whilst the identifier type box is defaulted to ‘Any’ it can be used to pre-allocate the source of the identifier. This allows the removal of ambiguity where a single identifier string is used in different resources—for instance both the CATH-PDB and CATH-HMM assignments use the same codes.
Figure 2.
The Gene3D search bar. This bar can be found at the top of all the Gene3D pages and is used to navigate the site. It consists of two main components—the query (A) and the filter (B)—that allow sophisticated data retrieval. Both components also consist of two inputs. (A) The first box describes the identifier type, with the default being any. Different resources often use identical identifier types to represent different proteins or protein families. As a result, the returned data can be ambiguous; users can restrict the identifier to a certain resource to remove ambiguity. The second box accepts the search term. (B) The filter allows the results to be restricted to particular subsets of the database. The first input is the filter type: at the moment ‘Genomes’, ‘GO Term’, ‘FunCat Category’ and ‘Affymetrix platform’. The second box accepts the filter term—for instance, ‘human’, ‘9606’ or ‘Mammalia’. (C) Possible terms for the query and the filter are shown as a drop-down list while the user types.
The second two boxes provide a filtering stage, limiting the results to a particular subset. As an example entering ‘genomes’ and ‘human’ will limit any returned results to the human genome; entering ‘genomes’ and ‘Mammalia’ will limit the results to completed mammalian genomes. To aid the user, the possible filter terms (i.e. human, man, 9606) are shown as a drop-down list that will refresh itself as the user types. Finally, ‘wildcard’ matches have also been added, allowing the retrieval of partially matched terms. So for instance, it is now simple to examine the annotation of members of the Ig Fold (CATH code 3.40.50) in humans and then to compare that with mammals in general. Some further example queries are detailed in Supplementary Data.
HMM and BLAST facilities
Whilst Gene3D contains a fairly comprehensive set of protein sequences we have also provided a facility for scanning user-provided sequences against the CATH HMM library with HMMER (31) and a BLAST (32) facility for identifying the most similar protein in the database. These facilities are designed for single sequence submissions, but we can also carry out genome-scale scans if requested.
DAS servers
The DAS servers have been recently re-implemented in ProServer 2 and we now provide four distinct services—for full paths and information on the servers please see the Supplementary Data. In summary, there are two Gene3D-specific servers, one providing the CATH HMM-assigned domain and one Gene3D protein cluster assignment for UniProt proteins. A third server provides the mapping of the CATH structural domains to UniProt, whilst a fourth provides the SPLIT 4.0 transmembrane region predictions. These servers are registered at the DAS registry and as a member of the BioSapiens network we will be actively working to create standards for improving the richness and display of the DAS content, as well as adding new servers.
Future Changes
We are constantly improving Gene3D and a comprehensive plan has been initiated to completely redesign the underlying hardware/software architecture. The result of these changes will be manifold. First, it will become much easier and quicker to make partial updates to the site and keep it up-to-date. Second, it will allow more sophisticated retrieval, analysis and display of data in the results tabs. For example, more dynamic family analysis pages will be developed and powerful predictive tools like PhyloTuner incorporated. It will also allow Gene3D to tightly bind itself to CATH releases, ensuring a minimum of delay before CATH PDB domains can be linked to genomic and functional data.
A second major change is that we will be receiving frequent updates for sequence similarity data and CATH HMM scan data from SIMAP allowing us to rapidly expand Gene3D and keep pace with the explosion of protein sequences coming out of sequencing projects. Furthermore, we hope to use the SIMAP Pfam HMM results to expand the Pfam assignments in Gene3D to cover all the sequences, rather than being restricted to those provided in Pfam-A.
Whilst there is already a large set of flat files available for download, it is only a fraction of the possible datasets that can be generated. We are always happy to provide these and to help other teams in utilizing the data for functional prediction, experimental targeting and evolutionary analyses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank EU Biosapiens Network and the Wellcome Trust for providing funding to create the Gene3D resource. We would also like to thank the CATH teams for their help, Juan Antonio Ranea for leading the development of Gene3D-based prediction methods and Thomas Rattei at SIMAP for generously providing the protein similarity data and hosting the CATH HMMs on the SIMAP BOINC system. Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust.
Conflict of interest statement. None declared.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 5.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 6.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman HM, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 8.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kersey P, Bower L, Morris L, Horne A, Petryszak R, Kanz C, Kanapin A, Das U, Michoud K, et al. Integr8 and genome reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee D, Grant A, Marsden RL, Orengo C. Identification and distribution of protein families in 120 completed genomes using Gene3D. Proteins. 2005;59:603–615. doi: 10.1002/prot.20409. [DOI] [PubMed] [Google Scholar]
- 11.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, et al. IntAct–open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marsden RL, Lewis TA, Orengo CA. Towards a comprehensive structural coverage of completed genomes: a structural genomics viewpoint. BMC Bioinformatics. 2007;8:86. doi: 10.1186/1471-2105-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn RD, Stalker JW, Jackson DK, Kulesha E, Clements J, Pettett R. ProServer: a simple, extensible Perl DAS server. Bioinformatics. 2007;23:1568–1570. doi: 10.1093/bioinformatics/btl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Güldener U, Münsterkötter M, Oesterheld M, Pagel P, Ruepp A, Mewes HW, Stümpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnikov N, Lilja P, et al. ArrayExpress – a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKusick VA. A Catalog of Human Genes and Genetic Disorders. 12th. Baltimore, Maryland: John Hopkins University Press; 1998. Mendelian inheritance in man. [Google Scholar]
- 22.Liu H, Hu ZZ, Zhang J, Wu C. BioThesaurus: a web-based thesaurus of protein and gene names. Bioinformatics. 2006;22:103–105. doi: 10.1093/bioinformatics/bti749. [DOI] [PubMed] [Google Scholar]
- 23.Martin AC. Mapping PDB chains to UniProtKB entries. Bioinformatics. 2005;21:4297–4301. doi: 10.1093/bioinformatics/bti694. [DOI] [PubMed] [Google Scholar]
- 24.Tress ML, Martelli PL, Frankish A, Reeves GA, Wesselink JJ, Yeats C, Olason PL, Albrecht M, Hegyi H, et al. The implications of alternative splicing in the ENCODE protein complement. Proc. Natl Acad. Sci. USA. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian W, Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? J. Mol. Biol. 2003;333:863–882. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- 26.Rost B. Enzyme function less conserved than anticipated. J. Mol. Biol. 2002;318:595–608. doi: 10.1016/S0022-2836(02)00016-5. [DOI] [PubMed] [Google Scholar]
- 27.Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;16:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 28.Rattei T, Arnold R, Tischler P, Lindner D, Stumpflen V, Mewes HW. SIMAP: the similarity matrix of proteins. Nucleic Acids Res. 2006;34:D252–D256. doi: 10.1093/nar/gkj106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 30.Ranea JA, Yeats C, Grant A, Orengo CA. Predicting protein function with hierarchical phylogenetic profiles: the Gene3D phylo-tuner method applied to eukaryotic genomes. PLoS Comput. Biol. 2007:e237. doi: 10.1371/journal.pcbi.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]