Abstract
Sequence similarity/database searching is a cornerstone of molecular biology. PairsDB is a database intended to make exploring protein sequences and their similarity relationships quick and easy. Behind PairsDB is a comprehensive collection of protein sequences and BLAST and PSI-BLAST alignments between them. Instead of running BLAST or PSI-BLAST individually on each request, results are retrieved instantaneously from a database of pre-computed alignments. Filtering options allow you to find a set of sequences satisfying a set of criteria—for example, all human proteins with solved structure and without transmembrane segments. PairsDB is continually updated and covers all sequences in Uniprot. The data is stored in a MySQL relational database. Data files will be made available for download at ftp://nic.funet.fi/pub/sci/molbio. PairsDB can also be accessed interactively at http://pairsdb.csc.fi. PairsDB data is a valuable platform to build various downstream automated analysis pipelines. For example, the graph of all-against-all similarity relationships is the starting point for clustering protein families, delineating domains, improving alignment accuracy by consistency measures, and defining orthologous genes. Moreover, query-anchored stacked sequence alignments, profiles and consensus sequences are useful in studies of sequence conservation patterns for clues about possible functional sites.
INTRODUCTION
Proteins are major constituents of all cells and central to our understanding of molecular biology. The past and present genome projects have provided us with an exponentially growing wealth of protein sequences from many diverse organisms. However, this wealth creates a new problem. The majority of protein sequences have not been studied experimentally in the laboratory and our first-hand knowledge about them is minimal. Inferring functions of protein sequences is a major challenge in bioinformatics.
The function of a novel protein sequence can often be inferred by studying similar protein sequences in other organisms. Just as whole organisms are related to each other by evolutionary descent, so are the proteins between organisms related by inheritance and mutation. Protein sequences in different organisms sharing an ancestral protein sequence are believed to fulfil the same function and are said to belong to the same protein family. Common ancestry between protein sequences is commonly established by studying protein similarity. During evolution, protein sequences accumulate mutations, but parts of the protein, which are central to its function will remain conserved due to selection. These conserved parts give rise to a detectable similarity between related sequences.
PairsDB facilitates the establishment of family relationships between all known protein sequences in order to assign functions to novel proteins and then to identify conserved parts in the protein sequences, which are essential for its function. Clustering methods to determine family relationships rely on all versus all comparison of all protein sequences with each other. For example, the CluSTr database (http://www.ebi.ac.uk/clustr/) offers an automatic classification of the the UniProt Knowledgebase based on Smith–Waterman alignments, SIMAP (http://boinc.bio.wzw.tum.de/boincsimap/) is a community project providing an all-against-all similarity matrix generated using the FASTA program, and CoGenT++ (http://cgg.ebi.ac.uk/cgg/cpp_sitemap.html) is a platform for computational research in comparative and functional genomics built upon an all-on-all similarity matrix for completely sequenced genomes. In addition to BLAST, PairsDB uses a highly sensitive searching method, i.e. PSI-BLAST (1), which does an incremental database search each time refining its search model. The method requires a lot of computational power and thus the computing was performed using the supercomputing environment at CSC (The Finnish IT Centre for Science). Given the considerable investment of resources, the results are made available to all. For example, PairsDB retrieves pre-computed PSI-BLAST search results in seconds, which take minutes to run individually (Table 1). If the database gained popularity, this could lead to significant savings of CPU cycles worldwide.
Table 1.
Wall-clock timings for query BRCA1_HUMAN
| Search method | E-value threshold | Source database | Matches | Seconds |
|---|---|---|---|---|
| BLAST | 1 | NRDB100 | 3343 | 7 |
| BLAST | 0.001 | NRDB100 | 1697 | 4 |
| BLAST | 1 | NRDB90 | 1604 | 1 |
| PSI-BLAST | 1 | NRDB100 | 3298 | 8 |
| PSI-BLAST | 0.001 | NRDB100 | 1697 | 4 |
| PSI-BLAST | 1 | NRDB40 | 705 | 1 |
MATERIALS AND METHODS
Building of PairsDB
PairsDB is a relational database of sequence relationships. It contains a comprehensive collection of protein sequences and BLAST and PSI-BLAST alignments between them. PairsDB is built in six steps: (i) collection of protein sequences, (ii) sequence annotation, (iii) redundancy removal at 90% identity, (iv) BLAST all-on-all alignment, (v) creation of non-redundant sequence subsets and (vi) PSI-BLAST all-on-all alignments. Details follow.
Collection of protein sequences
PairsDB is constructed by retrieving peptide sequences from a variety of publicly available databases. The current sequence data set of PairsDB was collected from the following datasets: UniProt (http://www.ebi.ac.uk/uniprot), PDB Protein Databank (http://www.pdb.org), ENSEMBL (http://www.ensembl.org) and RefSeq (http://www.ncbi.nlm.nih.gov/RefSeq). Cross-references to the source database are stored for each sequence entry in PairsDB. The peptide sequences are collated into a single input set. Identical sequences are merged into a single entry, which has a stable, unique identifier. The set of unique sequences is called NRDB (alias NRDB100).
Sequence annotations
Composition-biased regions (2) are detected by an in-house perl script written by G. Casari. Transmembrane segments are detected by tmhmm (Version 1.1) (3). Coiled coils are detected using Coils (Version 2.1) (4). Short repeats are identified using an in-house perl script. Data on domain family membership of a sequence entry is imported from ADDA (5), CATH (6), InterPro (7) and SCOP (8). Domain information is further mapped at the residue level to the representative sequences in the NRDB90 and NRDB40 sets described below. Hypothetical proteins, fragments and sequences from completely sequenced genomes are flagged.
Redundancy removal at 90% identity
The input set is pruned of highly redundant sequences using the nrdb90 method (9) as implemented by CD-HIT (10). This method begins by sorting all sequences by length in decreasing order. Starting from the longest sequence (representative sequence), the procedure removes all sequences from the set that align over their full length and are more than 90% identical to the selected sequence. The procedure then takes the second longest sequence and does the same. The procedure continues, until all sequences have been processed. The remaining set constitutes a set of representative sequences at 90% sequence identity, NRDB90. Because of the high similarity threshold, most alignments need not be calculated explicitly, but instead a fast tuple lookup algorithm is sufficient. The CD-HIT program was run with the following parameters: -M 2000 (memory/Mb) -n 5 (word length) -c 0.90 (sequence identity threshold) -d 40 (length of description line).
BLASTP all-on-all alignments
For all sequences in the representative set NRDB90, we run BLASTP in an all-on-all fashion. We use NCBI BLAST (Version 2.28) with the following parameters: -e 1.0 (maximum reported e-value) -z 65 000 000 (effective database size) -b 100 000 (alignments reported) -v 100 000 (hits reported) -F F (seg filtering off). Sequences submitted to BLAST are masked by composition bias, transmembrane regions, coiled coils and short repeats. The alignments are stored in a relational database.
Creation of RSDB
Using the alignment results from the previous steps, we produce a set of nested non-redundant sequence subsets for 80%, 70%, 60%, 50%, 40% and 30% sequence identity (11). These are called, synonymously, RSDBxx or NRDBxx, where xx is the level of sequence identity. The procedure works similarly to the creation of NRDB90, but percent identity is now retrieved from the BLAST results stored in the relational database.
PSI-BLAST all-on-all alignments
We run PSI-BLAST for all sequences in the representative set NRDB40 in an all-on-all fashion. We use NCBI PSI-BLAST (Version 2.28) program blastpgp with the following parameters: -j 10 (number of iterations) -e 1 (maximum e-value of reported hits) -h 0.001 (inclusion threshold) -v 100 000 (hits reported) -b 100 000 (alignments reported). Sequences submitted to PSI-BLAST are masked by composition bias, coiled coils and short repeats. The results are stored in a relational database.
Generation of alignments
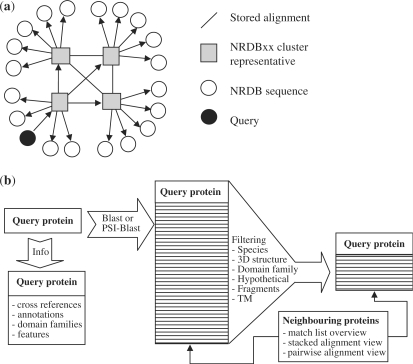
We compute the all-on-all alignment data in representative subsets of the whole sequence dataset. Sequences not in the representative subset are aligned only to the representatives (Figure 1a). All sequences are more than 90% identical to their representatives in the case of a BLAST search, and more than 40% identical in the case of a PSI-BLAST search. Using representative subsets saves search time and storage space quadratically and without loss of information (11). As NRDBxx clusters have a skewed size distribution, our scheme leads to huge savings in the largest protein families. The theoretical, and in our experience reasonable, assumption here is that alignments to a third party by sequences which are more than 90% or more than 40% identical to each other will be consistent. The alignment between two proteins of interest can be reconstructed on the fly by transitive alignment using their respective representatives as intermediates.
Figure 1.
(a) Data storage model and (b) Functionality of the PairsDB web server.
Interactive queries
The web server supports four types of query: (i) SeqInfo allows you to view all information about a particular protein sequence of interest from the PairsDB database; (ii) BLAST allows you to find BLAST matches for a sequence; (iii) PSI-BLAST allows you to find PSI-BLAST matches for a sequence; (iv) Seq Space Filter allows you to find a set of sequences satisfying a set of criteria. On each result page, shortcuts ‘I B P’ next to a match take you to the SeqInfo, BLAST, and PSI-BLAST views, respectively, of that sequence.
BLAST and PSI-BLAST results are shown as a match list (with graphical overview, similarity score and sequence description) and in stacked or pairwise alignment views (Figure 1b). The stacked alignment representation was popularized by HSSP (12) and is also used for example to generate profiles in PSI-BLAST (1). Stacked alignments show matching sequences underneath the query sequence, omitting insertions in the matching sequence. Pairwise alignments show the entire peptide sequence of both parties.
PSI-BLAST, especially, can produce match lists with thousands of neighbours. Therefore it is an important feature of PairsDB that the match list can be filtered based on sequence features, species, and family classifications, and truncated at an e-value threshold.
Programmatic access
In order to enable the integration of PairsDB to other applications and workflow tools such as Taverna (http://taverna.sourceforge.net/), we are currently developing a SOAP Web service interface for it. Further information including the description document in WSDL (Web service description language) will be available on the PairsDB web site.
Below, we describe two utility scripts, which use a local copy of the PairsDB database in MySQL. The scripts are available on request. The list of options in expand_pairsdb.pl is illustrated in Supplementary Figure 1b. PairsDB data tables are of two types: (i) attributes (features, flags) of individual sequences, and (ii) alignments between two sequences. Alignments are divided to two parts: the alignments/mappings of all sequences to the representative subset, and the alignments between all representatives (Figure 1a). Sequence attributes are used for filtering the match list, while a little additional computation is required to generate transitive alignments, query-anchored stacked alignments in various formats, profiles and consensus sequences.
PairsDB data covers only sequences already in Uniprot, PDB, Ensembl or Refseq. New sequences not yet in the source databases need to be mapped to a sequence known to PairsDB. A second utility script, called pairsdb_mapper.pl, identifies the nearest neighbour of a query sequence, which can be given as database identifier, accession number, amino acid sequence or domain range (Supplementary Figure 2). Multiple queries can be given in a fasta-file or list of fasta-files. The script uses a hash-code lookup to identify identical amino acid sequences, while a suffix array search recognizes identical fragments; if these fail to find a match, a BLAST run is performed. The user can substitute these mappings for the default PairsDB table in expand_pairsdb.pl.
Data updates
We currently update PairsDB 1–2 times per year. We are planning to move a weekly incremental update cycle for the BLAST all-on-all comparisons, with a less frequent update cycle for PSI-BLAST searches, which must start from scratch. The data is stored in a MySQL relational database. Data files will be made available for download at ftp://nic.funet.fi/pub/sci/molbio.
RESULTS
Our latest database contains 3.4 M unique protein sequences. After removing trivially similar sequences, we obtain 2.0 M and 1.1 M representative protein sequences in NRDB90 and NRDB40, respectively. The all versus all comparison yields 4.5 G and 2.3 G similarity relationships in the all-on-all comparison of NRDB90 by BLAST and of NRDB40 by PSI-BLAST, respectively.
The PairsDB web server allows these similarity relationships to be explored online and interactively (Figure 1b). Large-scale analyses should work on a local copy of the database.
To avoid excessively long and redundant match lists, we generally recommend searching against NRDB90 (default for BLAST results) or NRDB40 (default for PSI-BLAST results). These results are typically retrieved in a few seconds, compared to many minutes if the search is re-run with BLAST or PSI-BLAST (Table 1). Similarity searches are often done to infer the function of hypothetical proteins based on what is known about their nearest neighbours. For example, restricting the source database to PDB retrieves possible fold assignments for a query (using the ‘PSI/BLAST results expanded to NRDB100’ form on the PSI/BLAST query pages). Thus, you can see by just one click that the structural coverage of BRCA1_HUMAN spans an N-terminal ring domain and a C-terminal BRCT domain.
DISCUSSION
Molecular biologists are nowadays trained to do bioinformatics using web servers. PairsDB can substitute for BLAST or PSI-BLAST servers (such as NCBI's and EBI's) when the query sequence has been already deposited in a sequence database. Based on pre-computed results, our web server has a faster response time (Table 1). The Blink (BLAST link) service of NCBI Entrez has had a similar BLAST neighbour lookup since many years. An added value of PairsDB is that it can report more distant neighbours detected by PSI-BLAST and display query-anchored stacked alignments. The stacked alignment views are useful to get a picture of which residues are conserved in a family and thereby might be targeted in functional studies. Many domain family classifications also provide multiple sequence alignments [e.g. ADDA (13), CDD (14), PFAM (15)], but PairsDB has flexible filtering options to focus on matches to proteins of interest for a particular study. PairsDB shows the ‘raw’ similarity data without enforcing any particular classification of the sequences. This can actually be beneficial in allowing the user to critically evaluate electronically inferred functional annotations of suspect cases.
Computational biologists can build derivative tools exploiting PairsDB data. For example, a simple BLAST or PSI-BLAST search reveals only the radial distance of neighbouring sequences from the query sequence, whereas all-on-all similarity data reveals the topology of protein sequence space and clusters of biologically related sequences are seen more clearly (16–17). We have used the all-on-all similarity data to define protein families by a graph clustering method, ADDA. Considering all protein sequences for the clustering allows us to merge nearest neighbours hierarchically and to count exhaustively (5). A novel breed of sequence alignment methods take a library of pairwise alignments as input and use consistency criteria to suppress noise (18–22). We have developed methods based on this data that perform sensitive alignment between very distantly related protein sequences by taking into account intermediate sequences. Even when there is no direct alignment with PSI-BLAST, our method constructs a meaningful alignment (18–19). Furthermore, we have demonstrated how the data can be used to find functionally important parts in protein sequences that are conserved between all or a subset of members in a protein family (23–24). Finally, reciprocal best BLAST hits are conveniently extracted from PairsDB and can be used to determine orthologous genes in completely sequenced genomes in the hope of more accurate function assignment (25). We expect that the SOAP interface will lead to a proliferation of applications and web servers exploiting PairsDB data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
PairsDB integrates data from a variety of sources and we would like to thank the members of the UniProt, ENSEMBL, RefSeq, PDB teams for their efforts and their generous release of data to the public. The work was supported by the Academy of Finland (grant 1102273) and the EMBRACE project, which is funded by the European Commission within its FP6 Programme, under the thematic area ‘Life sciences, genomics and biotechnology for health’, contract number LHSG-CT-2004-512092. Funding to pay the Open Access publication charges for this article was provided by the Academy of Finland (grant 1102273).
Conflict of interest statement. None declared.
REFERENCES
- 1.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Promponas VJ, Enright AJ, Tsoka S, Kreil D, Leroy C, Hamodrakas S, Sander C, Ouzounis CA. CAST: an iterative algorithm for the complexity analysis of sequence tracts. Complexity analysis of sequence tracts. Bioinformatics. 2000;16:915–922. doi: 10.1093/bioinformatics/16.10.915. [DOI] [PubMed] [Google Scholar]
- 3.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 4.Lupas A, van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 5.Heger A, Holm L. Exhaustive enumeration of protein domain families. J. Mol. Biol. 2003;328:749–767. doi: 10.1016/s0022-2836(03)00269-9. [DOI] [PubMed] [Google Scholar]
- 6.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holm L, Sander C. Removing near-neighbour redundancy from large protein sequence collections. Bioinformatics. 1998;14:423–429. doi: 10.1093/bioinformatics/14.5.423. [DOI] [PubMed] [Google Scholar]
- 10.Li W, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 11.Park J, Holm L, Heger A, Chothia C. RSDB: representative protein sequence databases have high information content. Bioinformatics. 2000;16:458–464. doi: 10.1093/bioinformatics/16.5.458. [DOI] [PubMed] [Google Scholar]
- 12.Sander C, Schneider R. The HSSP database of protein structure-sequence alignments. Nucleic Acids Res. 1994;22:3597–3599. [PMC free article] [PubMed] [Google Scholar]
- 13.Heger A, Wilton CA, Sivakumar A, Holm L. ADDA: a domain database with global coverage of the protein universe. Nucleic Acids Res. 2005;33(Database issue):D188–D191. doi: 10.1093/nar/gki096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchler-Bauer A, Panchenko AR, Showmaker BA, Thiessen PA, Geser LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heger A, Holm L. Towards a covering set of protein family profiles. Prog. Biophys. Mol. Biol. 2000;73:321–337. doi: 10.1016/s0079-6107(00)00013-4. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Rost B. Domains, motifs and clusters in the protein universe. Curr. Opin. Chem. Biol. 2003;7:5–11. doi: 10.1016/s1367-5931(02)00003-0. [DOI] [PubMed] [Google Scholar]
- 18.Heger A, Lappe M, Holm L. Accurate detection of very sparse sequence motifs. J. Comp. Biol. 2004;11:843–857. doi: 10.1089/cmb.2004.11.843. [DOI] [PubMed] [Google Scholar]
- 19.Heger A, Mallick S, Wilton C, Holm L. The global trace graph, a novel paradigm for protein sequence database searching. Bioinformatics. 2007;23:2361–2367. doi: 10.1093/bioinformatics/btm358. [DOI] [PubMed] [Google Scholar]
- 20.Notredame C, Holm L, Higgins DG. COFFEE: an objective function for multiple sequence alignments. Bioinformatics. 1998;14:407–422. doi: 10.1093/bioinformatics/14.5.407. [DOI] [PubMed] [Google Scholar]
- 21.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for multiple sequence alignments. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 22.Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marttinen P, Corander J, Törönen P, Holm L. Bayesian search of functionally divergent protein subgroups and their function specific residues. Bioinformatics. 2006;22:2466–2474. doi: 10.1093/bioinformatics/btl411. [DOI] [PubMed] [Google Scholar]
- 24.Heger A, Holm L. Sensitive pattern discovery with ‘fuzzy’ alignments of distantly related proteins. Bioinformatics. 2003;19(Suppl. 1):i130–i137. doi: 10.1093/bioinformatics/btg1017. [DOI] [PubMed] [Google Scholar]
- 25.Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologs. Bioinformatics. 2003;19:1710–1711. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.