Abstract
Vaccines are among the most efficacious and cost-effective tools for reducing morbidity and mortality caused by infectious diseases. The vaccine investigation and online information network (VIOLIN) is a web-based central resource, allowing easy curation, comparison and analysis of vaccine-related research data across various human pathogens (e.g. Haemophilus influenzae, human immunodeficiency virus (HIV) and Plasmodium falciparum) of medical importance and across humans, other natural hosts and laboratory animals. Vaccine-related peer-reviewed literature data have been downloaded into the database from PubMed and are searchable through various literature search programs. Vaccine data are also annotated, edited and submitted to the database through a web-based interactive system that integrates efficient computational literature mining and accurate manual curation. Curated information includes general microbial pathogenesis and host protective immunity, vaccine preparation and characteristics, stimulated host responses after vaccination and protection efficacy after challenge. Vaccine-related pathogen and host genes are also annotated and available for searching through customized BLAST programs. All VIOLIN data are available for download in an eXtensible Markup Language (XML)-based data exchange format. VIOLIN is expected to become a centralized source of vaccine information and to provide investigators in basic and clinical sciences with curated data and bioinformatics tools for vaccine research and development. VIOLIN is publicly available at http://www.violinet.org
INTRODUCTION
Even in the face of major medical advances, infectious organisms remain a major source of morbidity and mortality worldwide. The World Health Organization estimates that infectious diseases were the cause of 14.7 million deaths in 2001, accounting for 26% of total global mortality (1). Since the introduction of Edward Jenner's cowpox-based vaccine against smallpox in 1796, vaccines have proven useful in their ability to stimulate the immune system and confer protection against infections by pathogenic microorganisms. As such, vaccines provide a safe, effective and cost-effective means to reduce the incidence of infectious diseases.
The most commonly developed types of vaccines include live attenuated vaccines, inactivated or ‘killed’ vaccines, toxoid vaccines and subunit vaccines. Live attenuated vaccines are derived from microbes that have been weakened by natural or genetically engineered mutations to a form that cannot cause disease. Inactivated vaccines are derived from whole viruses or bacteria that have been chemically or heat inactivated. Toxoid vaccines contain an inactivated toxin (toxoid) and are used to protect against toxins produced by bacteria. Subunit vaccines use one or more components of a disease-causing organism, rather than the whole organism to stimulate a protective immune response. Other types of vaccines currently in use or in development include conjugate, DNA and recombinant vector vaccines (2). Conjugate vaccines are comprised of proteins linked to the molecules of the outer coat of disease-causing bacteria. DNA vaccines utilize the genetic materials of a microbe to stimulate an immune response. Recombinant vector vaccines use modified viruses or bacteria to express genes that code for microbial antigens presented to cells of the host.
Vaccine research and development has undergone a renaissance in recent years. This is in part attributable to the cost-effectiveness of vaccines and advanced post-genomic technologies (3). However, although considerable progress has been made, vaccination against many medically important viruses (e.g. Human immunodeficiency virus), bacteria (e.g. Mycobacterium tuberculosis) or parasites (e.g. Plasmodium falciparum) has been unsuccessful for a variety of reasons, many related to the basic biology of the microbe and its interactions with the immune system. More extensive research is required in order to elucidate the pathogenesis and protective immune responses against these diseases (4).
Vaccine research and development continues to be actively pursued because of the need to control or eliminate remaining persistent pathogens. As the number of vaccine-related papers rises, it becomes increasingly challenging to identify, manage and annotate relevant articles without the availability of efficient literature mining and curation programs (5). There exists a marked need for a means to store, annotate, compare and analyze vaccine information across different pathogens and within different hosts. Although various vaccine databases (e.g. the Vaccine Page: http://www.vaccines.org/) exist that list commercialized vaccines and their usages, no publicly available central data repository are available to store and analyze research data concerning commercial vaccines and those vaccines under clinical trials or in early stages of research. Therefore, we have developed vaccine investigation and online information network (VIOLIN), a web-based database system designed to fill this need, i.e. to manually curate, store and permit analysis of published vaccine data (http://www.violinet.org).
SYSTEM AND DATABASE DESIGN
VIOLIN is implemented using a three-tier architecture built on two Dell Poweredge 2580 servers that run a Redhat Linux operating system (Redhat Enterprise Linux ES 4). Users submit database queries through the web. These queries are processed using PHP/SQL (middle-tier, application server based on Apache) against a MySQL (version 5.0) relational database (back-end, database server). The result of each query is then presented to the user through the web browser. This system is routinely backed up with two servers.
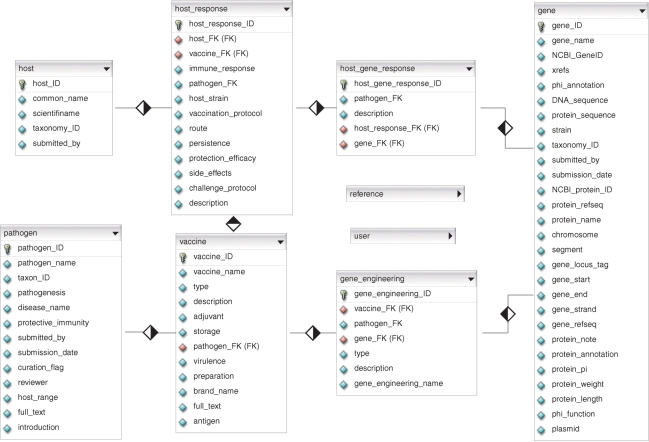
The VIOLIN database is populated by two primary sources of data, published vaccine literature downloaded from PubMed and manually curated vaccine data (Figure 1). The vaccine literature module includes basic literature information extracted from PubMed, such as article title, journal name, authors, year and medical subject headings (MeSH). Wherever possible, the PDF files of manuscripts are downloaded from PubMed. The PDF files are subject to text processing and are used for text mining in a program called Vaxpresso in VIOLIN. These files are very useful during internal curation; however, due to copyright and licensing restrictions, VIOLIN users may not be able to directly download them.
Figure 1.
Entity relationship diagram of the VIOLIN database. The details of table reference and user are not shown in this figure. The diamond represents a one-to-many (blank-to-solid) relationship between two tables.
VIOLIN presents information on the following topics organized as main database tables: pathogens, vaccines, vaccine-related genes including genetically modified pathogen genes and host responses to vaccines including specific host gene responses (Figure 1). The table detailing the pathogen includes general information about microbial pathogenesis, natural and experimental host ranges, and host protective immunity. Multiple vaccines are identified for each pathogen within the database. Each vaccine is assigned a unique accession number that serves as a central reference point in VIOLIN. In addition, each vaccine is characterized by a number of attributes such as vaccine type, preparation method, recommended storage conditions and gene manipulations. Vaccine candidates are often tested in multiple animal models using a variety of vaccination protocols. During these tests, the immunologic and physiologic responses of the animal are often characterized. Data from these tests are captured within the database and are found listed under the topics of host species (and strains or stocks, if applicable), vaccination protocol, persistence, side effects, host immune response, challenge protocol and observed efficacy. This information is necessary for understanding and comparing successful vaccine development strategies. The gene-engineering element describes and classifies the genetic engineering of pathogen genes utilized in each vaccine's development; these include recombinant protein preparation, gene mutation, DNA vaccine preparation and others. Host gene responses to each vaccine are also summarized in VIOLIN. Each gene entry contains several categories of information, including gene name, NCBI Gene database and/or other database IDs and DNA or protein sequences. The curated vaccine database emphasizes gene level information allowing a more specific analysis and comparison of vaccine research data.
Overall, the VIOLIN database design follows a logical and structured approach that facilitates efficient storage, curation, query and transfer of vaccine information. Based on this database, various query methods have been developed to search the vaccine literature and manually curate data. A web-based interactive literature mining and curation system is available for efficiently searching, curating and submitting vaccine data. Curated data can be exported using an eXtensible Markup Language (XML)-based data exchange format VIOLINML. These features comprise a comprehensive and integrative vaccine information repository and data mining system.
Vaccine literature download and search
All vaccine-related literature is obtained from PubMed by utilizing the search terms: ‘(pathogen name OR specific disease name) AND vaccine’. As of 30 October 2007, VIOLIN contains 24, 345 vaccine-related publications, including 24, 345 abstracts and 10, 317 full-text papers. These numbers will continue to increase as the database is updated every two months. The availability of these publications in the VIOLIN database allows development of more efficient vaccine literature mining tools than are possible through a standard PubMed search. The downloaded literature also provides a primary data source for vaccine curators to search and annotate structured vaccine data.
VIOLIN contains four integrated literature mining and search programs each with a different focus. These include: Litesearch, Vaxpresso, Vaxmesh and Vaxlert.
Litesearch is an advanced keyword- and category-based search for vaccine literature. Keywords can be searched from each of many subjects in a given publication including full text, abstract, article or journal title, author, year, issue and page numbers. Boolean search is supported. The output of a keywords search is comprised of a list of manuscripts contained within the VIOLIN literature database. Within this output page, the user can link to the results of an identical keyword search in other literature search programs.
Vaxpresso is a vaccine literature mining program powered by Textpresso, an open-source information retrieval system (http://www.textpresso.org) (Supplemental Figure 1) (6). Textpresso includes a natural language processing (NLP) program that splits papers into sentences and further to XML-tagged words or phrases classified using specifically designed categories of ontology. The ontology system, defined by Textpresso and other standard ontology resources, can be used to query information about specific categories or biological terms (e.g. gene) and their relationships (e.g. association or regulation). Vaxpresso uses the Textpresso NLP program to process all vaccine-related abstracts and available full text articles. Additionally, it provides a modified version of the user interface to retrieve and sort manuscripts with sentences containing requested keywords and ontology-based categories for selective pathogens. The results from keywords-based Litesearch and MeSH-based Vaxmesh searches can be linked from or to the Vaxpresso result page.
Vaxmesh is a vaccine literature browser based on MeSH (Supplemental Figure 2). MeSH is the controlled vocabulary of medical and scientific terms assigned by experts and used for indexing articles in PubMed (7). MeSH terminology provides a consistent approach to retrieve information that may use different terms to describe the same concepts. Vaxmesh enables users to locate articles using MeSH terms in a hierarchical MeSH tree structure. A specific MeSH term can also serve as a keyword with links to search results derived from other literature-searching programs.
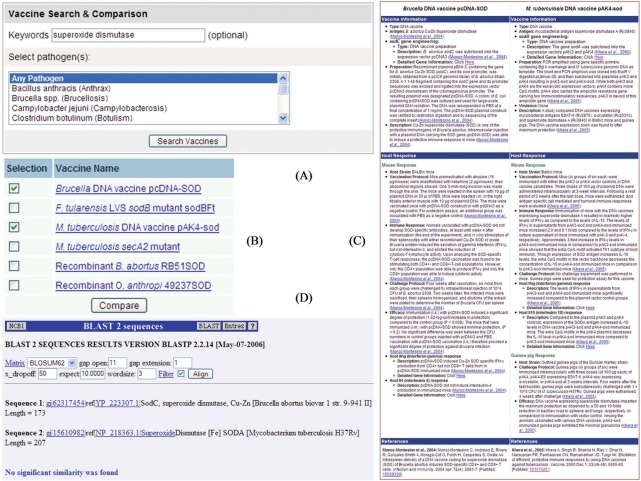
Figure 2.
Example of vaccine search and comparison. A keyword search of ‘superoxide dismutase’ (SOD) in Vaxquery (A) identified five vaccines curated in VIOLIN, including the Brucella abortus DNA vaccine expressing the gene sodC encoding for Cu/Zn superoxide dismutase and M. tuberculosis expressing the gene sodA encoding for Fe superoxide dismutase (B). DNA vaccines against either enzyme have been found to be effective against challenge infections by virulent B. abortus or M. tuberculosis strains (C). However, a VIOLIN BLAST-2-Sequences search did not find significant similarity between these two SOD protein sequences (D).
The strength of VIOLIN is its continued inclusion of recently published literature. Every two months, an internal VIOLIN program called Vaxlert extracts newly published vaccine literature related to targeted pathogens from PubMed, and then parses and stores it in the VIOLIN database. The Vaxlert website also displays new peer-reviewed papers published in the previous and current months. The new publication records are updated every hour. The users can access these publications even though they have not been stored in VIOLIN and manually curated. Vaxlert also includes an email alert service. Any subscribed user can specify the notification frequency (daily, weekly, bimonthly or monthly) and the keywords for a PubMed search. A daily Linux cron job checks the subscription database, searches for updates in PubMed, and sends notification to the users through Email (8). The email alert service also notifies data curators of newly published papers that may be appropriate for inclusion in VIOLIN.
ONLINE CURATION AND SUBMISSION
All information within the database originates from peer-reviewed literature and reliable websites. Much of the contents are curated internally, however, VIOLIN allows registered users to submit data for review and potential addition to the database. A web-based literature mining and curation system, Limix, is included in VIOLIN that permits efficient curation. The VIOLIN Limix system is an updated version of the Limix system originally developed in Brucella Bioinformatics Portal (BBP) for Brucella genome annotation (5). The VIOLIN Limix system includes two integrated components (panels) within one web screen, computational literature mining as well as manual curation and management (Supplemental Figure 3). The computational literature mining component contains the individual vaccine literature search programs described in the above section. Users can search for literature by going directly to PubMed, or access another webpage by entering a URL on the same Limix page. The manual curation and management component includes text fields in which curators can directly enter, edit and submit data. By integrating these components on a single webpage, a curator can perform computational text mining while simultaneously entering, editing and ultimately submitting data to the backend database.
At the time of submission, VIOLIN contained information for >200 vaccines or vaccine candidates against 18 pathogens (Table 1). These pathogens are selected based on the severity and impact of corresponding infectious diseases to the public, such as AIDS, malaria and tuberculosis. Many pathogens listed in VIOLIN are related to bioterrorism, including Bacillus anthracis and Francisella tularemia (9). The peer-reviewed papers (>480) have been curated manually in VIOLIN. New vaccines are continuously being added to the VIOLIN vaccine database. In order to track the numbers of curated vaccines and referenced papers, an internal program has been developed to update automatically the statistics and display them on the VIOLIN webpage at http://www.violinet.org/stat.php
Table 1.
Curated vaccine data in VIOLIN as of 30 October, 2007
| Pathogen (Disease) | No. of vaccines | No. of references |
|---|---|---|
| Bacillus anthracis (anthrax) | 14 | 29 |
| Brucella spp. (Brucellosis) | 19 | 48 |
| Campylobacter jejuni (Campylobacterosis) | 4 | 27 |
| Clostridium botulinum toxin (botulism) | 8 | 15 |
| Coxiella burnetii (Q fever) | 6 | 29 |
| Ebola virus (hemorrhagic fever) | 11 | 9 |
| Escherichia coli (Hemorrhagic colitis) | 15 | 21 |
| F. tularensis (tularemia) | 6 | 10 |
| H. influenzae (meningitis) | 20 | 44 |
| Hantavirus (pulmonary syndrome) | 4 | 8 |
| Human immunodeficiency virus (AIDS) | 13 | 25 |
| Lassa Fever virus (lass fever) | 9 | 7 |
| Marburg virus (hemorrhagic fever) | 7 | 7 |
| M. tuberculosis (Tuberculosis) | 17 | 30 |
| P. falciparum (Malaria) | 15 | 68 |
| Vaccinia virus (smallpox vaccine) | 12 | 28 |
| Venezuelan equine encephalitis virus (encephalitis) | 7 | 39 |
| Yersinia pestis (plague) | 24 | 36 |
| Total | 211 | 480 |
CURATED DATA QUERY AND DISPLAY
Vaxquery is a VIOLIN program that provides flexible and powerful means for querying and comparing curated vaccine data. Vaxquery supports three search formats, a vaccine search, a pathogen search and an advanced hierarchical data comparison. All three programs support keyword search in different categories using Boolean mode methodology. In the final output, the keywords are highlighted within the text for easier identification. If multiple categories are selected, the keywords searched are highlighted in different colors. When greater than one vaccine is found and selected in a vaccine search, Vaxquery displays information about the vaccines in parallel for ready comparison (Figure 2). The advanced hierarchical search and comparison program provides a hierarchical structure of the VIOLIN data and allows users to display selected vaccine information. Overall, these query and visualization approaches offer the advantages that they allow users to customize their search for vaccine-related information.
VACCINE-RELATED GENES AND BLAST ANALYSIS
Two groups of genes are emphasized in the VIOLIN database, pathogen genes used for vaccine development and host genes involved in protective immunity. Examples of vaccine-related pathogen gene components include recombinant pathogen proteins produced by bacteria or yeast, bacterial virulence genes that are mutated for preparation of live attenuated vaccines, and genes that are cloned and expressed in plasmids as components of DNA vaccines. Vaccine-related host gene components include protective immune defense factors such as interferon-gamma (IFN-γ) (10). Such genes have been curated from the web-based Limix vaccine submission system and are summarized in the VIOLIN component Vaxgen. During the Vaxgen data submission, a curator is capable of retrieving gene details from PHIDIAS, a program recently developed by the authors for study of pathogen–host interactions (11). Vaxgen also allows users to query and analyze vaccine-related genes. As of 30 October 2007, Vaxgen includes >120 vaccine-related pathogen or host genes.
DNA or protein sequences of all vaccine-related genes contained in Vaxgen have been included in customized BLAST libraries. This is significant since BLAST sequence similarity search programs are available for users to search genes that are similar to DNA or protein sequences. BLAST also allows users to search similar sequences against whole microbial genomes. The customized BLAST programs are designed to identify sequence similarity and specificity and to facilitate functional analysis of vaccine-related genes across different host species, strains and stocks (Figure 2D).
DATA TRANSFER AND DOWNLOAD
To facilitate data exchange and transfer, an XML-based data exchange format, VIOLINML, has been developed to represent vaccine data available in the VIOLIN database. VIOLINML allows for a portable, system-independent, machine- and human-readable representation of general vaccine information for individual pathogens. A user-friendly web page exists to describe the specifications of each VIOLINML element and attribute. XML uses simple text-based markup to describe the structure and semantics of ordered data, thereby allowing for standardized data formatting and interchange (12,13). The VIOLINML format has been defined by Document Type Definition (DTD) and XML Schema. All vaccine data stored in VIOLIN have been transferred into VIOLINML documents for public access.
DISCUSSION
VIOLIN is unique in that it is a web-based database system that focuses on research data mining, curation and analysis of vaccines of commercial use, or vaccine candidates in clinical trials or early stages of development. VIOLIN stores vaccine-related literature data downloaded from PubMed and provides various literature mining tools. These tools facilitate efficient identification and comparison of desired articles based on keywords and MeSH. They also permit searching for sentences in abstracts or full text that contain requested keywords and ontology-based categories. These advanced searching tools not only provide users powerful literature mining approaches, but also allow VIOLIN data curators to quickly find and annotate vaccine data from the literature. The database is powered by the Limix-based, interactive data submission and review system. This web-based program is a distributed curation system capable of involving external experts to support vaccine data curation efforts. Direct submissions from scientists keep the database comprehensive, updated and accurate. VIOLIN contains effective query and analysis tools for users to search curated data and analyze vaccine-related genes. Through its use, successful vaccine strategies are easily identified and may be utilized for designing future vaccine candidates.
VIOLIN will continue to be expanded and refined in the future. Information on additional pathogens and vaccines will be curated and included in the database. Additional bioinformatics programs are being developed within VIOLIN to facilitate vaccine research and development. For example, to aid in the design of future vaccines we are developing a component of VIOLIN that will predict vaccine target genes and epitopes based on genome sequences.
We anticipate that VIOLIN will become a timely and vital source of vaccine information and will provide investigators in the basic and clinical sciences with curated data and bioinformatics tools that will aid in the development of vaccines to fight infectious diseases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
Author Z.X. was supported by NIH-NIAID R21 grant (#1R21AI057875-01) to Y.H. and Startup Funding (Y.H.) from the University of Michigan. T.T. was supported by NIH T32 training grant #RR007008-30 at the Unit for Laboratory Animal Medicine at the University of Michigan. K.P.K, B.L.K., C.B.L. and E.A.O. were supported by the Undergraduate Research Opportunity Program (UROP) at the University of Michigan. Funding to pay the Open Access publication charges for this article was provided by the Startup Funding to Y.H. from the University of Michigan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Becker K, Hu Y, Biller-Andorno N. Infectious diseases – a global challenge. Int. J. Med. Microbiol. 2006;296:179–185. doi: 10.1016/j.ijmm.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.NIH-NIAID. Understanding Vaccines, What they are, How they Work. 2003. http://www.niaid.nih.gov/publications/vaccine/pdf/undvacc.pdf. November 08, 2007.
- 3.Almond JW. Vaccine renaissance. Nat. Rev. 2007;5:478–481. doi: 10.1038/nrmicro1702. [DOI] [PubMed] [Google Scholar]
- 4.NIAID. The Jordan Report: Accelerated Development of Vaccines 2007. 2007. http://www3.niaid.nih.gov/about/organization/dmid/PDF/Jordan2007.pdf. November 08, 2007.
- 5.Xiang Z, Zheng W, He Y. BBP: Brucella genome annotation with literature mining and curation. BMC Bioinformatics. 2006;7:347. doi: 10.1186/1471-2105-7-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lipscomb CE. Medical subject headings (MeSH) Bull. Med. Libr. Assoc. 2000;88:265–266. [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen R. 4th edn. Emeryville, CA: McGraw-Hill Osborne Media; 2000. Linux: The Complete Reference. [Google Scholar]
- 9.He Y, Rush HG, Liepman RS, Xiang Z, Colby LA. Pathobiology and management of laboratory rodents administered CDC category A agents. Comp. Med. 2007;57:18–32. [PubMed] [Google Scholar]
- 10.He Y, Vemulapalli R, Zeytun A, Schurig GG. Induction of specific cytotoxic lymphocytes in mice vaccinated with Brucella abortus RB51. Infect. Immun. 2001;69:5502–5508. doi: 10.1128/IAI.69.9.5502-5508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang Z, Tian Y, He Y. PHIDIAS: a pathogen–host interaction data integration and analysis system. Genome Biol. 2007;8:R150. doi: 10.1186/gb-2007-8-7-r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bray T, Paoli J, Sperberg-McQueen CM. Extensible Markup Language (XML) 1998. 1.0. Available at http://www.w3.org/TR/1998/REC-xml-19980210.html. November 08, 2007.
- 13.He Y, Vines RR, Wattam AR, Abramochkin GV, Dickerman AW, Eckart JD, Sobral BW. PIML: the Pathogen Information Markup Language. Bioinformatics. 2005;21:116–121. doi: 10.1093/bioinformatics/bth462. [DOI] [PubMed] [Google Scholar]