Abstract
The NONCODE database is an integrated knowledge database designed for the analysis of non-coding RNAs (ncRNAs). Since NONCODE was first released 3 years ago, the number of known ncRNAs has grown rapidly, and there is growing recognition that ncRNAs play important regulatory roles in most organisms. In the updated version of NONCODE (NONCODE v2.0), the number of collected ncRNAs has reached 206 226, including a wide range of microRNAs, Piwi-interacting RNAs and mRNA-like ncRNAs. The improvements brought to the database include not only new and updated ncRNA data sets, but also an incorporation of BLAST alignment search service and access through our custom UCSC Genome Browser. NONCODE can be found under http://www.noncode.org or http://noncode.bioinfo.org.cn.
INTRODUCTION
The considerable number of non-coding RNAs (ncRNAs) that has been detected in the past few years was largely unexpected (1–3). Although the functions of the many recently identified ncRNAs remain mostly unknown, increasing evidence stands in support of the notion that ncRNAs represent a diverse and important functional output of most genomes (4). NONCODE is an integrated knowledge database dedicated to ncRNAs. All ncRNAs in NONCODE were filtered automatically from GenBank (5) and the literature, and were then later manually curated. With the exception of rRNAs and tRNAs, all classes of reported ncRNAs are included. The aim of the database is to provide a platform that will facilitate both bioinformatic as well as experimental research. In addition to containing sequence data, NONCODE provides a user-friendly interface, a visualization platform and a convenient search option, allowing efficient recovery of sequences, regulatory elements in the flanking sequences, related publications and other information.
DATA COLLECTION AND ANNOTATION
Data collection and annotation for NONCODE v2.0 was carried out in a similar fashion as for version 1.0 and can be briefly described as follows: GenBank entries constituted the major source of NONCODE. We searched PubMed (6) with a list of ncRNA keywords, such as ‘ncRNA’, ‘snoRNA’, ‘snRNA’, ‘tmRNA’, ‘SRP RNA’, ‘gRNA’, etc., and thereafter consulted the literature matched with them and extracted more ncRNA keywords. The downloaded GenBank files (gbfiles) were then filtered using these keywords, and the filtered entries were subsequently confirmed by manual curation. For all obtained ncRNA records, basic information related to sequence, name, alias, length, ncRNA class, organism, references and accession number in GenBank were extracted and entered into the NONCODE database. Each ncRNA sequence was checked for redundancies using Perl scripts, and each cluster of redundant sequences was given a non-redundant NONCODE accession number (UniqID, i.e. unique ncRNA i.d.). In addition to the ‘traditional’ ncRNA classification system, NONCODE v1.0 introduced the alternative ‘process function class (PfClass)’ system based on the biological processes or functions in which an ncRNA is involved, and one or more of the 26 PfClasses were also assigned to all ncRNAs in NONCODE v2.0. Moreover, a subset of ncRNAs has been divided into nine additional categories according to whether they are gender- or tissue-specific or associated with tumors and diseases, etc. Where possible, NONCODE also provides additional annotations, such as information on function, cellular role, cellular location, chromosomal localization and splicing. The annotations and the genomic mapping information of the sequences rely on data provided in the original GenBank records, the FANTOM3 Database (2), the UCSC Genome Browser Database (7), or directly from the reference literature.
DATABASE CONTENT AND CLASSIFICATION
The purpose of the database is to serve the research community by organizing information concerning all types of ncRNAs (except tRNAs and rRNAs) from all groups of organisms. As of August 2007, the NONCODE database includes over 206 226 non-redundant sequences from 861 organisms. The significant growth in the amount of data, compared with the 5339 non-redundant sequences in the previous edition published in 2005, is primarily due to systematic identification of mRNA-like ncRNA transcripts (2) and the discovery of Piwi-interacting RNAs (piRNAs) through large-scale cDNA sequencing (1,3,8). Other novel ncRNAs, such as stem-bulge RNAs (sbRNAs) (9), snRNA-like RNAs (snlRNAs) (9) and a number of unclassified ncRNA transcripts were mainly obtained from our laboratory and other published literature (10–12). According to the traditional classification system, NONCODE v2.0 contains three novel classes of ncRNAs, the sbRNAs, the snlRNAs and the piRNAs, whereas the number of PfClasses is the same as in NONCODE v1.0 (i.e. 26), with sbRNAs and snlRNAs corresponding to the ‘Miscfunction_snm’ and piRNAs to ‘RNA-processing_cleavage’ PfClass.
DATABASE ACCESS
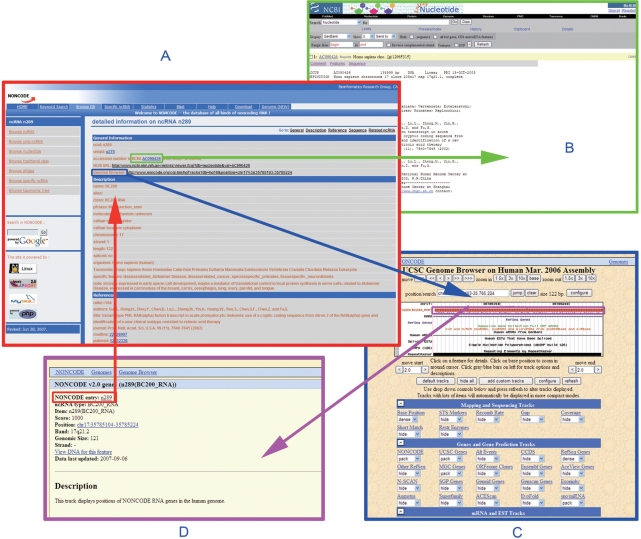
All sequences can be directly downloaded from the webpage. Sequences can be searched using accession numbers found in GenBank, name, traditional class, PfClass, organism and UniqID in NONCODE. In addition to access to NONCODE database records, search results are also linked to full GenBank entries (Figure 1). In the current version of the database, we also included the online BLAST service (NCBI wwwBLAST version 2.2.17) which allows sequence similarity searches against the entire NONCODE v2.0 database.
Figure 1.
Links between the NONCODE ncRNA annotations, the Genome Browser and NCBI. (A) The NONCODE database window with ncRNA annotations. (B) The corresponding NCBI annotation. (C) The corresponding Genome Browser window. (D) The link from Genome Browser to NONCODE.
In this updated version of NONCODE, a UCSC Genome Browser for NONCODE was constructed for Saccharomyces cerevisiae, Caenorhabditis elegans and Homo sapiens. NcRNA loci of these species may be viewed through the NONCODE track in the Genome Browser. Other common tracks concerning basic information on these species, such as mRNA genes, ESTs and so on, have also been retrieved from the UCSC Genome Browser Database. For the above three species, ncRNA entries in the NONCODE database can be directly linked to the Genome Browser; similarly, NONCODE ncRNA annotations may be accessed through the Genome Browser (Figure 1). The database can be accessed through the following URL: http://www.noncode.org/ or http://noncode.bioinfo.org.cn.
FUTURE DIRECTIONS
As new ncRNAs are being progressively discovered, we will continue to update the NONCODE database. Submissions of new ncRNAs are invited, and should be sent to noncode@ict.ac.cn. Within the coming year, we will continue to add Genome Browser services for other model organisms, such as mouse and fly. Given the increasing amount of ncRNA data and the emergence of ncRNA prediction software [e.g. QRNA (13), RNAz (14)], we will attempt to establish a service for ncRNA prediction based on the mentioned softwares and the information in the NONCODE database.
ACKNOWLEDGEMENTS
Sequence data were downloaded from NCBI GenBank (ftp://ftp.ncbi.nih.gov/genbank). The authors thank Lisa Caviglia for careful corrections. This work was supported by the National Key Basic Research & Development Program (973), under the Grant Nos. 2002CB713805 and 2003CB715907, the National Sciences Foundation of China, under Grant Nos. 30630040, 30570393 and 30600729, the Data Sharing Network of China Essential Medicine Science, under Grant No. 2005DKA32402. Funding to pay the Open Access publication charges for this article was provided by the National Sciences Foundation of China, under Grant No.30570393.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 4.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 5.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2007;35:D21–D25. doi: 10.1093/nar/gkl986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 9.Deng W, Zhu X, Skogerbo G, Zhao Y, Fu Z, Wang Y, He H, Cai L, Sun H, et al. Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis, and expression. Genome. Res. 2006;16:20–29. doi: 10.1101/gr.4139206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang ZP, Chen CJ, Zhou H, Li BB, Qu LH. A combined computational and experimental analysis of two families of snoRNA genes from Caenorhabditis elegans, revealing the expression and evolution pattern of snoRNAs in nematodes. Genomics. 2007;89:490–501. doi: 10.1016/j.ygeno.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Zemann A, op de Bekke A, Kiefmann M, Brosius J, Schmitz J. Evolution of small nucleolar RNAs in nematodes. Nucleic Acids Res. 2006;34:2676–2685. doi: 10.1093/nar/gkl359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rivas E, Eddy SR. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Washietl S, Hofacker IL, Stadler PF. Fast and reliable prediction of noncoding RNAs. Proc. Natl Acad. Sci. USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]