Abstract
The Structural Classification of Proteins (SCOP) database is a comprehensive ordering of all proteins of known structure, according to their evolutionary and structural relationships. The SCOP hierarchy comprises the following levels: Species, Protein, Family, Superfamily, Fold and Class. While keeping the original classification scheme intact, we have changed the production of SCOP in order to cope with a rapid growth of new structural data and to facilitate the discovery of new protein relationships. We describe ongoing developments and new features implemented in SCOP. A new update protocol supports batch classification of new protein structures by their detected relationships at Family and Superfamily levels in contrast to our previous sequential handling of new structural data by release date. We introduce pre-SCOP, a preview of the SCOP developmental version that enables earlier access to the information on new relationships. We also discuss the impact of worldwide Structural Genomics initiatives, which are producing new protein structures at an increasing rate, on the rates of discovery and growth of protein families and superfamilies. SCOP can be accessed at http://scop.mrc-lmb.cam.ac.uk/scop.
BACKGROUND
The Structural Classification of Proteins (SCOP) is a database of known structural and evolutionary relationships amongst proteins of known structure (1). By analogy with taxonomy, it has been created as a hierarchy of several obligatory levels. The fundamental unit of classification is a domain in the experimentally determined protein structure. Protein domains are grouped at different levels according to their sequence, structural and functional relationships. Proceeding from bottom to top, the SCOP hierarchy comprises the following levels: protein Species, representing a distinct protein sequence and its naturally occurring or artificially created variants; Protein, grouping together similar sequences of essentially the same functions that either originate from different biological species or represent different isoforms within the same organism; Family containing proteins with related sequences but typically distinct functions; and Superfamily bridging together protein families with common functional and structural features inferred to be from a common evolutionary ancestor. Near the root, the basis of classification is purely structural: structurally similar superfamilies with different characteristic features are grouped into Folds, which are further arranged into Classes based mainly on their secondary structure content and organization. The seven main classes in the latest release (1.73, forthcoming) contain 92 927 domains organized into 3464 families, 1777 superfamilies and 1086 folds. The SCOP domains correspond to 34 495 entries in the Protein Data Bank (PDB) (2). Statistics of the current and previous releases, summaries and full histories of changes and other information are available from the SCOP website (http://scop.mrc-lmb.cam.ac.uk/scop/) together with parseable files encoding all SCOP data (3). The sequences and structures of SCOP domains are available from the ASTRAL compendium (4), and hidden Markov models of SCOP domains are available from the SUPERFAMILY database (5).
Since the creation of SCOP in 1994, the number of known protein structures has grown more than 20-fold, whereas the numbers of SCOP folds, superfamilies and families have increased 4-fold, 5-fold and 7-fold, respectively. Besides an increased workload caused by the rapid data growth, processing these data for SCOP classification revealed more subtleties of protein relationships as well as new types of such relationships. It has become increasingly difficult to update the database while maintaining its original design. Accommodation of large numbers of new structures and their relationships within the SCOP hierarchy required some adjustments of the original classification scheme. In particular, there are large superfamilies, which continue to grow, accumulating many more new families and proteins. The division of these most populous superfamilies into families departed from the original SCOP scheme: their families consist of proteins of very similar structures that may or may not have a significant global sequence similarity. The proteins in these families are presumably more closely related to each other than to proteins in other more structurally divergent families.
A large proportion of new structures come from worldwide Structural Genomics initiatives, which are producing them at an increasing rate (6,7). Generally these structures are functionally uncharacterized which complicates their classifications at the Protein and Superfamily levels. Therefore, the initial classifications of such structures may be provisional. Discoveries of new relationships may either confirm these classifications or revise them. Other complications for a hierarchical classification come from the discoveries of probable remote homologies between superfamilies of distinct protein folds and the non-trivial structural relationships within sequence families (8,9).
While keeping the original classification scheme intact, we have changed the production of SCOP in order to cope with the growing amount of the new structural data and to facilitate the discovery of new distant relationships. Here we describe ongoing developments and new features implemented in the SCOP database and changes introduced to the SCOP update protocol. We also analyze the classification of new structures targeted by the structural genomics initiatives and discuss the impact of these initiatives on the rates of discovery and growth of protein families and superfamilies.
NEW SCOP UPDATE PROTOCOL
We have introduced significant changes to the SCOP update protocol since our previous report (10). These include a new data management system that supports batch analysis of new structures and their relationships and that also makes use of a relational database. We use a new protocol to produce SCOP that adds steps of fully automatic classification and pre-classification of protein structures. Following these changes, future SCOP releases will attempt to include important new content from the PDB archive by prioritizing the representative structures of members of new protein families, superfamilies and folds, although this will compromise their completeness. SCOP release 1.71 is the last release guaranteed to provide a complete survey of all PDB entries released before a certain date.
The relationships in SCOP are established by expert analysis of sequence, structural and functional similarities amongst proteins with known structure. Typically, the protein domains at the bottom levels of the classification hierarchy share significant sequence similarity that together with their common functional features suggests a common evolutionary origin. The protein relationships at the Superfamily level (and in some families) are more distant, but some of them can be detected with more advanced profile searches. Taking into account these organizational principles, we have developed a new update pre-classification protocol to optimize production and accelerate classification of new protein structures in SCOP. The methodology includes consecutive steps of sequence clustering and database searches using a combination of search methods. The similarity thresholds for these methods are set to less restrictive values than defaults, resulting in less stringent matches. This approach is justified because these borderline results will be subjected to manual inspection. It can suggest additional ‘true’ distant relationships, which otherwise would be missed. Database searches are performed using a selected non-redundant sequence set of representative update structures. The outputs of the database-search programs are analyzed and compared automatically and provisional classification is made for those representative proteins where a sequence match indicates a possible relationship to SCOP domains. While the new pre-classification protocol is entirely based on sequence comparisons, the analysis of structural similarities and final classification of the protein structures in the database will continue to rely on the SCOP authors’ knowledge and expertise.
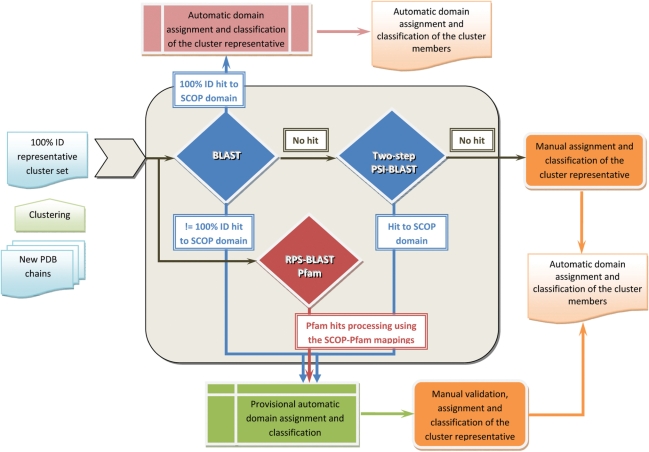
The workflow of the update protocol is shown in detail in Figure 1. The BLAST search allows detection of close homologs, which usually belong to the same SCOP family, whereas the two-step PSI-BLAST (11) and RPS-BLAST searches are used to identify similarities indicative of a more distant relationship (at the Superfamily level). Where the results of PSI-BLAST and RPS-BLAST methods overlap, they provide a consensus pre-classification that has proved to be reliable. In addition, each of these methods detects unique matches, which assist the final classification. For example, such a unique match suggested the relationship of an uncharacterized protein AF0060 from Archaeoglobus fulgidus (12) to the recently discovered superfamily of all-alpha NTP pyrophosphohydrolases with a potential ‘house cleaning’ function (13). This relationship was detected only by RPS-BLAST through Pfam families PF03819 and PF08761, both being linked to this SCOP superfamily. Manual inspection of the AF0060 structure confirmed the superfamily assignment, but also revealed its distinct features, including the subunit fold, tetrameric biological unit and active site architecture. Therefore we classify AF0060 to a new family of this superfamily. This example underlines intrinsic differences between the sequence-based annotations and the structural classification. For example, Pfam assigns AF0060 to the MazG-like family (PF03819), to which it shares a local sequence similarity, including the conserved metal ion-binding motif. The SCOP classification does not support this family assignment but suggests that AF0060 has a more distant relationship to the MazG-like proteins at the Superfamily level.
Figure 1.
Workflow of the SCOP update protocol. The update sequence set of new unclassified structures is derived from the PDB SEQRES record. Disordered regions at the termini are masked. The update sequences are clustered using a threshold of 100% identity and 95% coverage for the inclusion of protein sequence into the cluster set. The resulting clusters are used to select a representative sequence set. This dataset is used as a primary input to the pre-classification pipeline. The representative cluster set is first compared using BLAST against itself and a database of non-redundant representative ASTRAL sequences for SCOP domains. This step allows detection of close homologs, usually members of the same SCOP family. Representative sequences without significant sequence match (E-value = 0.001) are further used for two-step PSI-BLAST searches. In the first step, a position-specific scoring matrix (PSSM) is generated by searching the NCBI non-redundant protein database. The resulting PSSM is saved after ten PSI-BLAST iterations or less if the program converges. In the second step, each saved PSSM is used to scan databases of representative ASTRAL and update sequences. In addition, the representative cluster set of unclassified proteins is submitted for RPS-BLAST search against a database of Pfam profiles. The resulting matches are then compared with the matches of pre-computed large-scale comparisons of SCOP domains and Pfam families. A provisional SCOP classification assignment is made for those proteins with a matching region in Pfam that has given a hit to SCOP domain. The results of both RPS-BLAST and PSI-BLAST are used to identify relationships between more distant homologs that are likely to be members of the same SCOP superfamily. Update proteins that are identical or nearly identical to domains classified in the current SCOP release or in the SCOP developmental version are classified automatically. The remaining proteins with and without provisional classification are curated manually.
The computed sequence comparisons and the automatic provisional classifications of the update structures are imported into a MySQL relational database together with the SCOP classification hierarchy and detailed information on the PDB entries. All data are stored in a relational table format that offers the ability to capture all necessary information for a given protein structure such as PDB coordinate and sequence data, protein source and NCBI taxonomy (14), relationships to other proteins within the database and relationships to external sequence resources such as Pfam (15) and UniProt (16). Having the data available in the relational database allows complex queries and extensive analysis of the protein relationships. The database schema is flexible and it can readily accommodate third-party resources or new extensions of different types of automatic and manual annotations. It also allows continuous data flow inputs and hence synchronization with the PDB releases.
The new SCOP update system supports analysis of unclassified protein structures by the detected relationships amongst themselves and to already classified structures in contrast to our previous sequential handling of new structural data by release date. Thus, the manual curation of the representative update structures is carried out incrementally by analyzing the proteins’ Family and Superfamily relationships. Priority is given to representatives of new sequence families, in particular those targeted by structural genomics. This allows a rapid accumulation of new SCOP families, superfamilies and folds. Related structures with identical and nearly identical sequences to representative domains classified in the latest official SCOP release or in the SCOP developmental version are classified automatically. Possible structural diversity amongst these proteins usually arises from segment swapping, presence of chameleon sequences or functional conformational changes. A comprehensive annotation of these will be provided in the SISYPHUS database, a companion to the SCOP database (9).
PRE-SCOP—A PREVIEW OF THE SCOP DEVELOPMENTAL VERSION
In addition to the official SCOP build, starting with the release 1.71, we provide a preview of the SCOP developmental version (pre-SCOP, http://www.mrc-lmb.cam.ac.uk/agm/pre-scop/) that shows a snapshot of the ongoing classification update. Pre-SCOP is built using the classification data from the current SCOP release and data from the SCOP update. Pre-SCOP can be updated more frequently than the official SCOP release, as the curation of the pre-classified data progresses. This enables earlier access to the information on new structural relationships. SCOP users can request a higher priority to be given to the classification of a particular recent structure of general interest in PDB to be added to pre-SCOP.
At present there is no reclassification of the current SCOP entries in pre-SCOP. These entries retain the same ‘stable’ identifiers as in the latest SCOP release. All new nodes are assigned provisional six-digit identifiers counting down from 999 999. These identifiers are unique for a given release of pre-SCOP but are not stable across updates. The pre-SCOP Web interface allows browsing the classification hierarchy in a similar manner as the official SCOP site. A search engine can be used to find entries matching any identifiers or other text. When browsing pre-SCOP, all new folds are listed at the top of each Class page. In all other levels of the SCOP hierarchy, the new entries are listed at the bottom of the parent node page. Some functionalities of the official SCOP web site such as molecular viewer, expand and shrink options, and links to domain sequences are not available in pre-SCOP. New features unique to pre-SCOP include graphical display of the sequence–structure mappings hyperlinked to the SCOP, PDB and UniProt entries, available at the Domain level. Pre-SCOP also lists the provisional classifications derived from the automatic sequence comparisons. These are displayed only for the entries assigned to a given family at the family page.
NEW ADDITIONS AND CLASSIFICATION REFINEMENT
Changes in SCOP domain boundary definitions
Previously, SCOP domains that span an entire PDB chain did not have explicit beginning and end boundaries. These single domains may contain artificial sequences such as purification tags and other parts of cloning constructs. They may include unstructured parts of neighboring domains at one or both termini. These artifacts can result in significant sequence matches between unrelated SCOP domains. Starting with this release, we have introduced explicit begin–end boundaries for each new domain in SCOP and will gradually extend this to older entries. Thus, all protein domains in SCOP, with the exception of those that are artificially designed, will represent a distinct region of protein structure that has been derived from a natural protein sequence.
Integrated taxonomy
The SCOP unique identifiers (sunid) denote a unique node in the classification hierarchy and therefore do not allow retrieval of proteins or protein domains from a given organism. The SCOP species name does not always coincide with the name of the protein's biological source but also may represent a distinct sequence variant (isoform) of the same functional protein in a given biological species. For example, human hemoglobin β-chain and its embryonic variants are classified as separate SCOP species. Therefore all SCOP domains from a given organism cannot always be retrieved by the species name. In order to provide a consistent taxonomic presentation of SCOP entries, we attributed the NCBI TaxIDs to the Species nodes. These relationships are provided as a parseable file.
Curated relationships to sequence databases
We have extended the scope of the curated data to SCOP–Pfam relationships and mappings of SCOP domains on UniProt sequences. Relationships between SCOP nodes and Pfam families are rather complex due to genuine design differences and technical underpinnings of these databases. The curated relationships between SCOP and Pfam families are currently shown mainly in the comment field at Family level. Similarly, many comments at Species level include curated UniProt domain boundaries of SCOP domains mapped to the corresponding sequences. Future SCOP releases will provide this information in a parseable format.
FUTURE DEVELOPMENTS
The new developments in SCOP assure a step to the future transitions we are planning to introduce in the database. An ongoing project aims to define a reference SCOP sequence that represents a set of identical or nearly identical proteins with known structure and corresponds to a natural sequence as deposited in the public databases. A large portion of the valuable information in SCOP is stored in the form of free-text description. We are currently working on converting these free-text comments into more structured and machine-readable format.
A major enterprise is the planned redefinition (and renaming) of the Fold level of the SCOP hierarchy to enable the grouping of superfamilies on more than one basis. This would allow the classifications of many more known relationships in SCOP while preserving its basic hierarchy.
SCOP AND REMEDIATED PDB
Remediation of the PDB archive resulted in a number of changes directly affecting the SCOP data, which have been ‘remediated’ in response. The reassignment of the PDB chain identifiers caused a corresponding reassignment in the affected SCOP domains, changing their sid identifiers. These identifiers, derived from the PDB filename and chain identifiers, are ‘short names’ for SCOP domains, conventionally used by bioinformatics databases and tools to link to the SCOP pages. We would like to advise that the SCOP unique identifiers (sunid) for these domains remain unchanged and will provide a stable reference to the remediated entries in SCOP. We also would like to caution that there are changes in some PDB sequences that may affect the associated ASTRAL data.
IMPACT OF STRUCTURAL GENOMICS ON THE DISCOVERY OF PROTEIN RELATIONSHIPS IN SCOP
We investigated the impact of structural genomics on the rates of discovery and growth of protein families and superfamilies. Using the new update procedure, we collected and classified the structures determined by worldwide Structural Genomics initiatives and related structures from structural biology projects. In our analysis, we focused on the two central levels of SCOP hierarchy, Family and Superfamily, specifying the probable near and far evolutionary relationships, respectively. Because the SCOP hierarchy is obligatory, each protein structure is assigned to a family and a superfamily, regardless of whether there is a related structure at the corresponding levels or not. If there is another protein in a family, it becomes a ‘true’ family; otherwise it is a ‘singleton’ family. Similarly, a ‘true’ superfamily must contain two or more families, whereas a ‘singleton’ superfamily consists of a single family.
There are classification caveats for functionally uncharacterized proteins. Those of similar sequences are usually grouped at Family level, but, when their functions are established, they may be reclassified at the lower, Protein level. Also, the lack of functional evidence hinders the discovery of Superfamily relationships for such protein families, so that it is not always achievable at the time of creation of new families in SCOP. If there is no such relationship discovered, a new ‘singleton’ superfamily is created. This classification can be revised during a subsequent database update, when new evidence appears. For example, the hypothetical protein PA1492 (17), initially classified in its own superfamily in release 1.69, was reclassified in the next release 1.71 in the N-(deoxy)ribosyltransferase-like superfamily. This superfamily also unified the functionally and structurally characterized N-deoxyribosyltransferase and ADP-ribosyl cyclase-like families, which structural and mechanistic similarities remained unrecognized for a decade since the unraveling of their structures. PA1942 has a very similar putative active site and is predicted by SCOP to have a deoxyribosyltransferase activity.
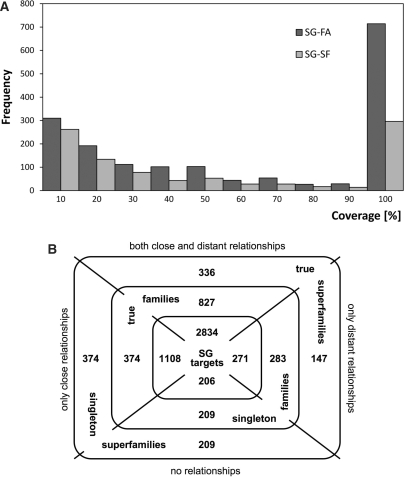
Hereafter, we refer to a SCOP family (superfamily) that contains a (domain of) structural genomics (SG) target as a SG-family (SG-superfamily, respectively). There were 1693 SG-families and 957 SG-superfamilies populated with domains from 4198 SG targets in PDB (released up to June 2007). For most of these domains, we found relationships to other proteins classified in SCOP at one or both of these two levels. For a small fraction (∼5%) of SG targets, we found no close or distant homolog. Only distant homologs were identified for <7% of SG targets. There was a nominal contribution to a small fraction (30%) of SG-families, where SG domains were neither the first nor the second member of the family and <30% of its content. About 30% of the SG-families were singletons. Nearly three-fifths of these singleton families belong to ‘true’ SG-superfamilies, and only 42% were a single member of superfamily (Figure 2).
Figure 2.
Statistics of SCOP classification of SG targets. (A) Numbers of SG-families and SG-superfamilies by fraction of SG domains in them. (B) Division of SG-families in ‘true’ and ‘singleton’ families, their SG target contents and their distribution in ‘true’ and ‘singleton’ superfamilies. Note that different parts of the same SG target can be classified into different families and that a ‘true’ superfamily can contain both ‘true’ and ‘singleton’ families.
Nearly half of the SG domains, at the time of their release, represented a new SCOP family. We observed a general trend, wherein the release of the first representative structure of a family was followed shortly by the release of a related structure determined by a different group of authors. In a few cases, more than one structure had been determined independently for the same protein.
A significant increase in the number of structurally characterized families facilitated the discovery of new relationships at the Superfamily level. More than half of families in ‘true’ SG-superfamilies contain SG domains. About half of SG domains defined a new family and hence contributed to the discovery of a new relationship. Analysis of the distribution of protein families characterized by structural genomics has confirmed a dominant role of the largest known superfamilies, which have grown further in the numbers of constituent families. There are other superfamilies, which have grown large rather unexpectedly (Table 1). The evolutionary success of these ‘new rich’ superfamilies is probably due to the presence of unusual conserved and, presumably, functionally important features in their folds. Interestingly, the first several structures of ‘metagenomic’ targets from environmental samples, selected as representative of novel sequence families (18), all belong to these ‘new rich’ superfamilies, mostly to the dimeric α + β barrel superfamily in SCOP.
Table 1.
Selected SG-superfamilies largely populated with SG-families
| SCOP Superfamily | Number of SG-FA | Coverage by SG-FA (%) | Representative structure |
|---|---|---|---|
| PUA domain-like | 12 | 100 | 1wmm |
| NagB/RpiA/CoA transferase-like | 7 | 100 | 2g40 |
| Alpha/beta knot | 6 | 100 | 1mxi |
| Ribokinase-like | 5 | 100 | 1ub0 |
| AhpD-like | 4 | 100 | 2cwq |
| ITPase-like | 3 | 100 | 1vp2 |
| RmlC-like cupins | 20 | 87 | 2atf |
| Dimeric α + β barrel | 18 | 78 | 1mwq |
| Bet v1-like | 6 | 67 | 1xfs |
| NTF2-like | 10 | 54 | 1tp6 |
CONCLUDING REMARKS
SCOP was created over a decade ago, when remote homology between proteins was mainly deduced from structural analysis. Since its creation, the tree-like classification has continuously evolved with the increasing amount of structural data. The recent advances in sequencing technologies resulted in a wealth of sequence data and the progress in profile-based database searches allowed the inference of new distant relationships from sequence analysis. While remaining focused on the structure-guided discovery of new evolutionary relationships at Superfamily level, SCOP now relies on the integrated sequence resources in the classification of new structures and refinement of the existing classifications.
The growth of structurally characterized protein families facilitated the discovery of new protein relationships in SCOP. Expansion of the existing protein superfamilies has provided additional structural and functional insights for their constituent members. The identification of new relationships has benefited from the apparent redundancy of joint structural biology and structural genomics efforts. Structural comparisons have revealed many examples of non-trivial protein relationships, suggesting that significant structural variations within sequence families are more common than were thought before.
ACKNOWLEDGEMENTS
This work was supported in part by the MRC strategic grant G0100305. A.A. and D.H. thank Prof. Sir Alan Fersht for the continuing financial support. J.-M.C. and S.E.B. are supported by NIH grant R01-GM073109. J.-M.C. is supported by the US Department of Energy under contract DE-AC02-05CH11231. Funding to pay the Open Access publication charges for this article was provided by the MRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 2.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo Conte L, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res. 2002;30:264–267. doi: 10.1093/nar/30.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandonia J-M, Hon G, Walker NS, Lo Conte L, Koehl P, Levitt M, Brenner SE. The ASTRAL compendium in 2004. Nucleic Acids Res. 2004;32:D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson D, Madera M, Vogel C, Chothia C, Gough J. The SUPERFAMILY database in 2007: families and functions. Nucleic Acids Res. 2007;35:D308–D313. doi: 10.1093/nar/gkl910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandonia J-M, Brenner SE. The impact of Structural Genomics: expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 7.Levitt M. Growth of novel protein structural data. Proc. Natl Acad. Sci. USA. 2007;104:3183–3188. doi: 10.1073/pnas.0611678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreeva A, Murzin AG. Evolution of protein fold in the presence of functional constraints. Curr. Opin. Struct. Biol. 2006;16:399–408. doi: 10.1016/j.sbi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Andreeva A, Prlic A, Hubbard TJP, Murzin AG. SISYPHUS – structural alignments for proteins with non-trivial relationships. Nucleic Acids Res. 2007;35:D253–D259. doi: 10.1093/nar/gkl746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreeva A, Howorth D, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nocek B, Xu X, Savchenko A, Edwards A, Joachimiak A. 2007. PDB ID: 2P06 Crystal structure of a predicted coding region AF_0060 from Archaeoglobus fulgidus DSM 4304. 10.2210/pdb2p06/pdb. [Google Scholar]
- 13.Moroz OV, Murzin AG, Makarova KS, Koonin EV, Wilson KS, Galperin MY. Dimeric dUTPases, HisE, and MazG belong to a new superfamily of all-α NTP pyrophosphohydrolases with potential “house-cleaning” functions. J. Mol. Biol. 2005;347:243–255. doi: 10.1016/j.jmb.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 14.Wheeler DL, Chappey C, Lash AE, Leipe DD, Madden TL, Schuler GD, Tatusova TA, Rapp BA. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2000;28:10–14. doi: 10.1093/nar/28.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2007;35:D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong A, Xu X, Liu Y, Zhang R, Savchenko A, Edwards A. 2004. PDB ID: 1T1J Crystal Structure of Conserved Hypothetical Protein PA1492 from Pseudomonas aeruginosa. 10.2210/pdb1t1j/pdb. [Google Scholar]
- 18.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, et al. The Sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]