Abstract
The Bacillus cereus group of bacteria is an important group including mammalian and insect pathogens, such as B. anthracis, the anthrax bacterium, B. thuringiensis, used as a biological pesticide and B. cereus, often involved in food poisoning incidents. To characterize the population structure and epidemiology of these bacteria, five separate multilocus sequence typing (MLST) schemes have been developed, which makes results difficult to compare. Therefore, we have developed a database that compiles and integrates MLST data from all five schemes for the B. cereus group, accessible at http://mlstoslo.uio.no/. Supertree techniques were used to combine the phylogenetic information from analysis of all schemes and datasets, in order to produce an integrated view of the B. cereus group population. The database currently contains strain information and sequence data for 1029 isolates and 26 housekeeping gene fragments, which can be searched by keywords, MLST scheme, or sequence similarity. Supertrees can be browsed according to various criteria such as species, isolate source, or genetic distance, and subtrees containing strains of interest can be extracted. Besides analysis of the available data, the user has the possibility to enter her/his own sequences and compare them to the database and/or include them into the supertree reconstructions.
INTRODUCTION
Multilocus sequence typing (MLST) is a tool that is widely used for phylogenetic typing of bacteria. MLST is based on polymerase chain reaction (PCR) amplification and sequencing of internal fragments of usually seven essential or housekeeping genes spread around the bacterial chromosome. The genetic relatedness among isolates is then determined by comparison of the nucleotide sequence types (1,2). MLST is thus a method that is unambiguous and truly portable among laboratories. Since the initial development of this technique for Neisseria meningitidis in 1998, MLST schemes have been developed for about 30 species including some of the most important bacterial pathogens, e.g. Streptococcus pneumoniae, Streptococcus pyogenes, Haemophilus influenzae, Staphylococcus aureus, Campylobacter jejuni, Enterococcus faecium, Burkholderia pseudomallei, Escherichia coli, Salmonella enterica and the Bacillus cereus group (see (1) for a recent review). These MLST schemes have been used successfully to explore the population structure of bacteria, to study the evolution of their virulence properties, to identify antibiotic-resistant strains and epidemic clones, and for epidemiological surveillance.
The B. cereus group includes bacterial species that are of medical and/or economic importance, such as B. anthracis, an obligate mammalian pathogen causing the lethal disease anthrax, B. cereus, an opportunistic human pathogen involved in food-poisoning incidents and contaminations in hospitals, B. thuringiensis, an insect pathogen and one of the world's most widely used biopesticide and B. weihenstephanensis, a cold-tolerant species known for contaminating dairies. These species are genetically very closely related and may be considered as one species based on genetic and genomic evidence (3–5). Unlike other bacterial species that are typed using a single MLST scheme, five separate schemes have been developed for the B. cereus group, based on different sets of genes and isolates (5–10). The Priest scheme (8) is currently the most widely used. Studies with the various schemes have independently indicated that the B. cereus group population is divided into three main phylogenetic clusters and that species are usually intermixed within the groups. One cluster contains the monomorphic B. anthracis isolates and a number of B. cereus and B. thuringiensis strains, many of which are from clinical sources. A second heterogeneous cluster includes B. cereus and B. thuringiensis isolates from various origins, while cold-tolerant B. weihenstephanensis and B. cereus isolates belong to the third group. The separate MLST analyses have also revealed that the B. cereus group population is weakly clonal overall due to numerous clinical and virulent isolates emerging from different phylogenetic positions (5–8,11–14), with the exception of the ‘cold-tolerant’ cluster that seems to exhibit a panmictic (or sexual) population structure, i.e. with frequent genetic exchanges between strains (9).
Despite the overall congruence between the various MLST studies, the use of separate schemes with no gene overlap and very little strain overlap has produced a confusing situation and makes the results difficult to compare directly. Therefore, we recently proposed a combined scheme based on genes taken from three of the four schemes available by then and for which we created a web-based database accessible at the University of Oslo's MLST server, http://mlstoslo.uio.no/ (5). Here, in order to provide the B. cereus group research community with a common MLST resource, we have developed on the same website a database, SuperCAT, that compiles and integrates MLST data from all the published B. cereus group schemes. In addition, we applied supertree reconstruction methods to build an integrated view of the B. cereus group population and phylogeny. Below we describe the content and main features of the new database as well as the process of supertree building.
DATABASE CONTENT AND IMPLEMENTATION
The SuperCAT database provides information, sequence and phylogenetic data for all bacterial isolates that have been typed using any of the five published MLST schemes for the B. cereus group (Table 1). Strain information, when known, includes isolate description, source and geographical location of isolation, and the scheme(s) used for typing. The sequence data include the nucleotide sequences of the MLST loci examined in a given strain. SuperCAT also contains the phylogenetic supertree of the B. cereus group reconstructed by combining the sequence data from all five schemes, as well as supertrees built for individual schemes. Information and sequences for isolates typed by the Priest and Tourasse–Helgason schemes were retrieved from the databases devoted to these schemes at http://pubmlst.org/bcereus and http://mlstoslo.uio.no/, respectively. MLST data for additional strains not available in the pubmlst.org repository (strains from (15) are missing therein) and for the Helgason, Ko, and Candelon–Sorokin schemes were taken from the published literature and the Genbank nucleotide sequence database (Table 1). In addition, sequences of all MLST loci were extracted from the complete genomes of the 21 sequenced B. cereus group strains available in Genbank. Altogether, SuperCAT currently contains data for 1029 isolates and 26 gene fragments from 25 different genes. However, since most strains have been typed using only 6 or 7 of the 26 loci, about one-third of the complete set of sequences are included. The 26 loci, only available for the completely sequenced strains, sum up to 10 619 bp. All these genes are located on the chromosome, thus the database provides no information about extrachromosomal plasmids even though most of the strains do carry one or several small and/or large plasmids. Unlike scheme-specific MLST databases, SuperCAT does not contain allele and sequence type (ST) numbers. Since isolates in SuperCAT have been typed by different subsets of loci, complete allelic profiles are unavailable and therefore STs cannot be assigned for most strains, except the fully sequenced ones.
Table 1.
The five MLST schemes designed for typing bacteria of the B. cereus group
| Scheme | Genes | Total sequence length (bp) | Total number of isolatese | Used in (references) |
|---|---|---|---|---|
| Helgason | adk, ccpA, ftsA, glpT, pyre, recF and sucC | 2 938 | 120 | (6,12,46) |
| Candelon–Sorokina, c | clpC, dinB, gdpD, panC, purF and yhfL | 2 850 | 149 | (9,10) |
| Kob, c | gyrB, mbl, mdh, mutS, pycA(1) and rpoB | 2 002 | 65 | (7) |
| Priesta, b | glpF, gmk, ilvD, pta, purH, pycA(2) and tpi | 2 829 | 721 | (8,11,13–15,46–48) |
| Tourasse–Helgasona, b, d | adk, ccpA, glpF, glpT, panC, pta and pycA(2) | 2 658 | 172 | (5) |
aSpecific databases for the Priest and Tourasse–Helgason schemes are accessible at http://pubmlst.org/bcereus/ and http://mlstoslo.uio.no/, respectively. A BLAST database for the Candelon–Sorokin scheme is available at http://spock.jouy.inra.fr/cgi-bin/bacilliMLSopen.cgi.
bWhile the Tourasse–Helgason and Priest schemes use the same gene fragment for the pycA gene, the Ko scheme is based on a different and non-overlapping gene region.
cThe B. cereus group-specific transcriptional regulator plcR was originally included in the Candelon–Sorokin and Ko schemes. However, plcR follows a phylogeny different from the other MLST loci (7,10) and is no longer used for MLST; therefore, it is not included in SuperCAT.
dThe Tourasse–Helgason scheme is a combined scheme based on 3 genes from the Helgason scheme (adk, ccpA, and glpT), 3 genes from the Priest scheme (glpF, pta and pycA(2)), and the panC gene from the Candelon–Sorokin scheme.
eIncluding strains with fully sequenced genomes.
SuperCAT is built as a relational database using the PostgreSQL management system, and data are accessible through a graphical web interface. User queries and results pages are processed and created on-the-fly via a highly modified version of the mlstdbNet software (16) written in PERL and based on the DataBase Interface (DBI) and Common Gateway Interface (CGI) modules. The database is implemented on a Linux Apache web server maintained through the facilities and support provided by the Norwegian EMBnet node. Some large supertree computations are run on a Linux supercomputer at the University of Oslo. The ATV (A Tree Viewer) Java applet is used for phylogenetic tree display (17). ATV notably supports horizontal and vertical zooming capabilities that are suitable for browsing large trees. The Jalview editor Java applet is also implemented in SuperCAT for advanced multiple sequence alignment display (18).
SUPERTREE RECONSTRUCTION
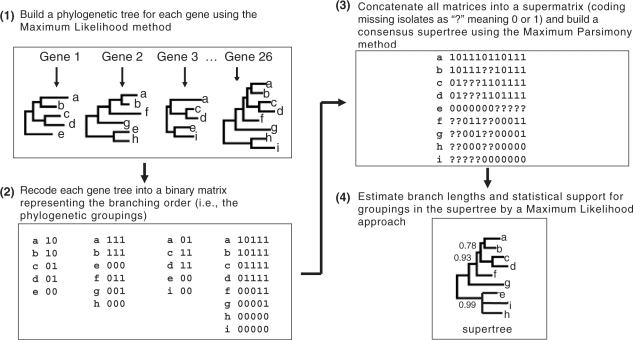
Supertree techniques allow to combine the phylogenetic information from different datasets into a common phylogenetic tree and several studies have shown that meaningful supertrees can be obtained even when taxon overlap is very sparse (see (19,20) for reviews). Supertree analysis has thus become increasingly popular for taking advantage and combining the massive amount of sequence data available in public databases for reconstructing large-scale organismal phylogenies with the ultimate goal of building the tree of life (21–24). In this study, the 21 B. cereus group strains that have been completely sequenced, and for which the sequences at all 26 MLST loci are thus available, can be used to join all five schemes and provide the strain overlap necessary for supertree analysis. The global B. cereus group supertree, containing 1029 isolates, was reconstructed according to the widely used matrix representation by parsimony (MRP) procedure (Figure 1; (19,25,26)). Scheme-specific supertrees were also reconstructed for each of the five MLST schemes by the same technique. Briefly, a phylogenetic tree is built for every gene separately by the maximum likelihood method with the PHYML_aLRT program (27). Then, each gene tree is recoded into a binary matrix representing the branching order (i.e. the phylogenetic groupings) following standard MRP coding using the SuperMRP.pl script (28). All gene tree matrices are concatenated into a supermatrix, in which isolates missing from a particular tree are coded using the ‘?’ character representing unknown data. In this supermatrix, the sequence of 0's, 1's and ?'s defines the branching profile of a strain. Closely related strains have similar branching profiles. Supertrees are then generated from the supermatrix by the maximum parsimony technique using the program MIX from the PHYLIP package (29) run with default parameters. The maximum parsimony step infers the trees that would require the minimum number of changes between the branching profiles of all isolates, where the unknown characters can take any of the two possible states 0 or 1 (they are not treated as missing gaps). As many trees were equally parsimonious, the final supertree was taken as the strict consensus of all parsimony trees with the CONSENSE program of PHYLIP. In order to obtain branch lengths that are proportional to the amount of nucleotide changes, we added an additional step in which branch lengths and statistical support for groupings are estimated from the concatenated sequences by the maximum likelihood method employing approximate likelihood-ratio tests (aLRTs) for branches using PHYML_aLRT with Shimodaira-Hasegawa-like support values (27,30). aLRTs provide a fast way of testing branch support without requirement of multiple replicates like traditional bootstrap procedures. The Felsenstein-1984 nucleotide substitution model supplemented with a gamma distribution (F84+Γ) was used in maximum likelihood computations for individual gene trees and the supertree (31). This model allows for unequal base frequencies, transition/transversion rate bias, and gamma-distributed substitution rate variation among sites. It was empirically chosen as a consensus from exploratory model testing using ModelTest (32,33), which indicated that models including these three factors were most appropriate for the MLST loci studied, although models for individual loci differed slightly. Note that the maximum likelihood technique also allows for uneven rates of nucleotide substitution between strains, which allows to accommodate slow- and fast-evolving isolates. To reduce the size of the binary supermatrix and speed up computations, individual gene trees and the supertree were built using only one representative from a set of strains having identical sequences. The remaining identical isolates were graphically added to the tree afterwards when drawing the final supertree.
Figure 1.
Schematic overview of the B. cereus group supertree reconstruction procedure using Matrix Representation by Parsimony (MRP). See text for details.
It should be noted that the global 1029-strain supertree retains the phylogenetic signals from the individual schemes and contains the three main clusters of the B. cereus group population described in the section ‘Introduction’. The integrated SuperCAT system may also allow to infer new relationships between strains that were analyzed with different gene sets. Even though the 26 loci sequences are available for only 21 isolates, they apparently provide enough overlap information for building the main branches of the supertree. These 21 isolates cover all three clusters, although the majority of them are B. anthracis strains or clinical strains closely related to B. anthracis due to the focus of genome sequencing projects, making the part of the supertree containing these isolates likely to be more accurate than the rest of the tree. Furthermore, 111 other isolates have been typed by 10 genes or more, providing additional overlap (see the ‘Gene Distribution’ page). Although about two-thirds of the sequence data are missing overall, it has been shown for other organisms that relevant supertrees could be reconstructed with datasets containing more than 90% of missing data, especially when the characters that are present are informative (20,22,23,34). Empirical and simulation studies have indicated that this behavior may be due to the fact that the characters which are present are more important for the tree-building process than those which are absent (see (20,34) and references therein). Precise within-cluster groupings may contain more uncertainty, as indicated by the large number of unresolved multifurcations in the B. cereus group supertree. Finally, it is also worth mentioning that the branching orders of the scheme-specific MRP supertrees are highly correlated to those of the published trees built with concatenated sequences and other phylogenetic algorithms.
DATA ACCESS AND MANIPULATION
The complete list of isolates included in SuperCAT (currently 1029) with strain description, source and country of origin is available at the database home page. By default all isolates in the database are used in the analysis tools provided, but the user can select strains of interest by keywords, MLST scheme, entering a list of strain identifiers, or choosing isolates individually via checkboxes. All subsequent analyses will be based on the selected strain subset and their loci. The keyword search will look for matches in any of the strain, description, source, location and scheme fields. Complex keyword queries with several logical operators can be formulated in the ‘advanced search’ page. Note that many isolates were referred by alternative names in different MLST schemes and publications, therefore synonyms have been included in the strain descriptions that allow a particular isolate to be looked up using any of its alternative identifiers. A sequence search is also possible using BLASTN (35), in order to select isolates that have allele sequences identical to user-entered query sequences.
Throughout SuperCAT, clicking on a strain name will pop up an isolate-specific window showing all relevant information and giving access to the nucleotide sequences of individual loci for that isolate. Detailed information about the MLST schemes (e.g. loci names and lengths, genomic coordinates, literature references) and their overlap, the distribution of available loci among the isolates, and the supertree reconstruction procedure can be obtained by clicking the relevant links in the header line present at the top of every page.
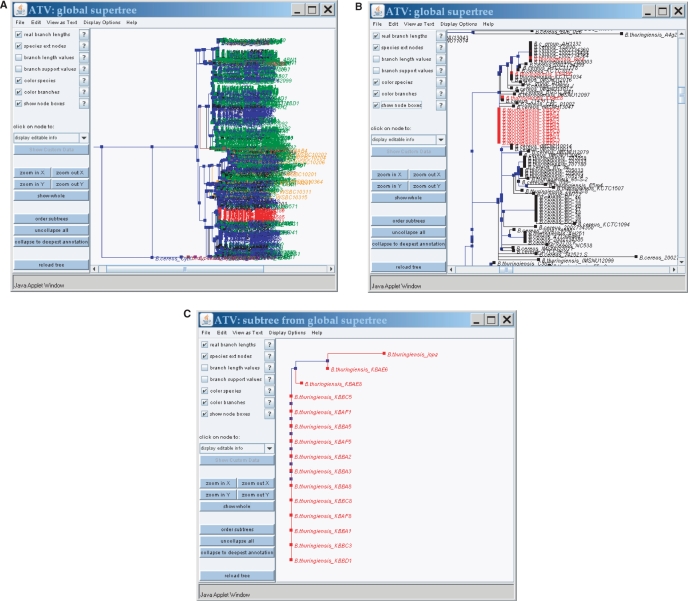
Apart from the basic functions for selecting and accessing strain information and sequence data for all five B. cereus group MLST schemes, the main features of SuperCAT relate to the manipulation of the supertrees constructed by the MRP approach. The global supertree based on the combination of all five B. cereus group MLST schemes as well as the five scheme-specific supertrees can be browsed according to various user-chosen criteria (Figure 2). Isolates in the supertrees can be colored by species or source of isolation. It is also possible to specifically mark in red the current subset of strains that has been selected by the user and to extract from the supertrees the subtree containing only those isolates. In the case of the multi-scheme supertree highlighting of the strains can be based on genetic distance. With this option the user can mark on and/or extract from the tree the isolates that are genetically closely related to strains of her/his choice. The user can either select strains that share one or several identical allele sequences with her/his query isolate(s) or that are at a specified genetic distance. Distances between isolates are computed by summing up the lengths of the branches (in average number of nucleotide substitutions per site) connecting the isolates in the supertree (known as patristic distances; (36)). The genetic relatedness search functions are also available in a separate page for the user to find closely related isolates without tree manipulation. SuperCAT allows to compare the scheme-specific MLST supertrees with each other and with the global supertree by using the subset of isolates that are common to all selected schemes. Common isolates can either be highlighted in red or be extracted as subtrees from each supertree, which can be used for comparing the positions of the common strains in the various MLST trees. For all supertree-related options, detailed tree navigation can be achieved using the various functions in the ATV tree window when the trees are displayed (17).
Figure 2.
Examples of supertree browsing and manipulation in SuperCAT. A, supertree colored by species; B, specific highlighting of user-selected strains (in red); C, extracted subtree containing only the strains highlighted in B. Trees are displayed using ATV (17).
Besides the manipulation of the precomputed supertrees, SuperCAT offers the user the possibility to compute new supertrees by MRP using any combination of strains, schemes and genes. Supertree computations may be extremely time consuming, ranging from a few minutes to 2–3 days with the complete database. Users are therefore requested to enter their e-mail addresses and will receive a notification containing a link to the results page when the supertree is ready. Note that when building a supertree for a user-selected subset of strains, the computation will first include all database isolates. A subtree containing only the user-selected isolates will then be extracted from the supertree of all strains. Although more time consuming, this strategy allows: (i) to avoid sampling artefacts as phylogenies built with different isolate sets may vary and (ii) to obtain relationships even if the selected isolates have been typed using non-overlapping gene sets, as the supertree of all isolates can always be built owing to the completely sequenced strains that are common to all schemes.
Another main feature of the SuperCAT database is that the user can enter her/his own private sequences and conduct several sequence analyses (Figure 3). These analyses include: (a) building new supertrees containing user isolates and sequences; (b) finding database isolates having sequences most similar to the user's query sequences using an on-line BLASTN (35) service; and (c) aligning user sequences to database genes using the multiple sequence alignment program CLUSTALW (37). For the last option, the Jalview editor (18) is provided for advanced multiple alignment display. All user-entered data must be in FASTA format and can be either copied and pasted into the query forms or uploaded from text files stored locally on the user's computer.
Figure 3.
Examples of query results in SuperCAT. A, multi-scheme BLAST search with sequence alignment; B, multi-scheme genetic search showing the list of isolates sharing one or more sequences with a query strain; C, multiple sequence alignment using Jalview (18).
All strain information data, sequences and phylogenetic trees, including user-made supertrees and extracted subtrees, can be saved and downloaded freely from the database. Users wishing to have their MLST data included as part of the SuperCAT release (and/or the Tourasse–Helgason scheme-specific database) are welcome to contact N.J.T. or A.-B.K. at the e-mail addresses given on the Oslo MLST server front page.
DISCUSSION AND FUTURE DEVELOPMENTS
SuperCAT is a newly created database devoted to the B. cereus group of bacteria whose main objectives are to provide a common MLST repository and means for building a comprehensive genetic analysis of the group that has been typed by five separate schemes. The database is publicly and freely available at http://mlstoslo.uio.no/, along with the database specific for the combined scheme of (5). We plan to update SuperCAT quarterly.
Future developments of the database may deal with refining the supertree-building procedure. In particular, a new improved method has recently been developed for taking into account both nucleotide substitutions and recombination events in phylogenies, as part of the ClonalFrame software, which has been applied to the B. cereus group and the Priest scheme (38). It would therefore be of interest to examine the suitability of ClonalFrame in the supertree context. It is also tempting to extend the supertree analysis beyond MLST data, by incorporating the phylogenies obtained previously from large-scale multilocus enzyme electrophoresis (MLEE; (3,39–41)) and amplified fragment length polymorphism (AFLP; (42–45)) studies. The MRP framework is ideal since it allows to integrate trees that can be built from different methods and data. As MLEE, AFLP and MLST have different levels of resolution, one can hope that combining them might provide an even more robust supertree for the B. cereus group.
ACKNOWLEDGEMENTS
We thank George Magklaras, The Biotechnology Center of Oslo and The Norwegian EMBnet node, University of Oslo, Oslo, Norway, for technical assistance and maintenance of the web server facilities. We also thank Erlendur Helgason, Section for Fish Health, National Veterinary Institute, Oslo, Norway, for helpful discussions. Funding to pay the Open Access publication charge was provided by by the Norwegian Consortium for Advanced Microbial Sciences and Technologies (CAMST) platform.
Conflict of interest statement. None declared.
REFERENCES
- 1.Maiden MC. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 2.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helgason E, Økstad OA, Caugant DA, Johansen HA, Fouet A, Mock M, Hegna I, Kolsto AB. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis–one species on the basis of genetic evidence. Appl. Environ. Microbiol. 2000;66:2627–2630. doi: 10.1128/aem.66.6.2627-2630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasko DA, Altherr MR, Han CS, Ravel J. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol. Rev. 2005;29:303–329. doi: 10.1016/j.femsre.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Tourasse NJ, Helgason E, Økstad OA, Hegna IK, Kolstø AB. The Bacillus cereus group: novel aspects of population structure and genome dynamics. J. Appl. Microbiol. 2006;101:579–593. doi: 10.1111/j.1365-2672.2006.03087.x. [DOI] [PubMed] [Google Scholar]
- 6.Helgason E, Tourasse NJ, Meisal R, Caugant DA, Kolstø AB. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 2004;70:191–201. doi: 10.1128/AEM.70.1.191-201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko KS, Kim JW, Kim JM, Kim W, Chung SI, Kim IJ, Kook YH. Population structure of the Bacillus cereus group as determined by sequence analysis of six housekeeping genes and the plcR Gene. Infect. Immun. 2004;72:5253–5261. doi: 10.1128/IAI.72.9.5253-5261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Priest FG, Barker M, Baillie LW, Holmes EC, Maiden MC. Population structure and evolution of the Bacillus cereus group. J. Bacteriol. 2004;186:7959–7970. doi: 10.1128/JB.186.23.7959-7970.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorokin A, Candelon B, Guilloux K, Galleron N, Wackerow-Kouzova N, Ehrlich SD, Bourguet D, Sanchis V. Multiple-locus sequence typing analysis of Bacillus cereus and Bacillus thuringiensis reveals separate clustering and a distinct population structure of psychrotrophic strains. Appl. Environ. Microbiol. 2006;72:1569–1578. doi: 10.1128/AEM.72.2.1569-1578.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Candelon B, Guilloux K, Ehrlich SD, Sorokin A. Two distinct types of rRNA operons in the Bacillus cereus group. Microbiology. 2004;150:601–611. doi: 10.1099/mic.0.26870-0. [DOI] [PubMed] [Google Scholar]
- 11.Barker M, Thakker B, Priest FG. Multilocus sequence typing reveals that Bacillus cereus strains isolated from clinical infections have distinct phylogenetic origins. FEMS Microbiol. Lett. 2005;245:179–184. doi: 10.1016/j.femsle.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Ehling-Schulz M, Svensson B, Guinebretiere MH, Lindback T, Andersson M, Schulz A, Fricker M, Christiansson A, Granum PE, et al. Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology. 2005;151:183–197. doi: 10.1099/mic.0.27607-0. [DOI] [PubMed] [Google Scholar]
- 13.Vassileva M, Torii K, Oshimoto M, Okamoto A, Agata N, Yamada K, Hasegawa T, Ohta M. Phylogenetic analysis of Bacillus cereus isolates from severe systemic infections using multilocus sequence typing scheme. Microbiol. Immunol. 2006;50:743–749. doi: 10.1111/j.1348-0421.2006.tb03847.x. [DOI] [PubMed] [Google Scholar]
- 14.Vassileva M, Torii K, Oshimoto M, Okamoto A, Agata N, Yamada K, Hasegawa T, Ohta M. A new phylogenetic cluster of cereulide-producing Bacillus cereus strains. J. Clin. Microbiol. 2007;45:1274–1277. doi: 10.1128/JCM.02224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim K, Cheon E, Wheeler KE, Youn Y, Leighton TJ, Park C, Kim W, Chung SI. Determination of the most closely related Bacillus isolates to Bacillus anthracis by multilocus sequence typing. Yale J. Biol. Med. 2005;78:1–14. [PMC free article] [PubMed] [Google Scholar]
- 16.Jolley KA, Chan MS, Maiden MC. mlstdbNet–distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics. 2004;5:86. doi: 10.1186/1471-2105-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zmasek CM, Eddy SR. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics. 2001;17:383–384. doi: 10.1093/bioinformatics/17.4.383. [DOI] [PubMed] [Google Scholar]
- 18.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 19.Bininda-Emonds OR. The evolution of supertrees. Trends Ecol. Evol. 2004;19:315–322. doi: 10.1016/j.tree.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 20.de Queiroz A, Gatesy J. The supermatrix approach to systematics. Trends Ecol. Evol. 2007;22:34–41. doi: 10.1016/j.tree.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Daubin V, Gouy M, Perriere G. A phylogenomic approach to bacterial phylogeny: evidence of a core of genes sharing a common history. Genome Res. 2002;12:1080–1090. doi: 10.1101/gr.187002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMahon MM, Sanderson MJ. Phylogenetic supermatrix analysis of GenBank sequences from 2228 papilionoid legumes. Syst. Biol. 2006;55:818–836. doi: 10.1080/10635150600999150. [DOI] [PubMed] [Google Scholar]
- 23.Driskell AC, Ane C, Burleigh JG, McMahon MM, O’Meara BC, Sanderson MJ. Prospects for building the tree of life from large sequence databases. Science. 2004;306:1172–1174. doi: 10.1126/science.1102036. [DOI] [PubMed] [Google Scholar]
- 24.Salamin N, Hodkinson TR, Savolainen V. Building supertrees: an empirical assessment using the grass family (Poaceae) Syst. Biol. 2002;51:136–150. doi: 10.1080/106351502753475916. [DOI] [PubMed] [Google Scholar]
- 25.Bininda-Emonds OR. Supertree construction in the genomic age. Methods Enzymol. 2005;395:745–757. doi: 10.1016/S0076-6879(05)95038-6. [DOI] [PubMed] [Google Scholar]
- 26.Bininda-Emonds OR, Sanderson MJ. Assessment of the accuracy of matrix representation with parsimony analysis supertree construction. Syst. Biol. 2001;50:565–579. [PubMed] [Google Scholar]
- 27.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 28.Bininda-Emonds OR, Beck RM, Purvis A. Getting to the roots of matrix representation. Syst. Biol. 2005;54:668–672. doi: 10.1080/10635150590947113. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. Seattle: University of Washington; 2006. [Google Scholar]
- 30.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J, Churchill GA. A Hidden Markov Model approach to variation among sites in rate of evolution. Mol. Biol. Evol. 1996;13:93–104. doi: 10.1093/oxfordjournals.molbev.a025575. [DOI] [PubMed] [Google Scholar]
- 32.Posada D. ModelTest Server: a web-based tool for the statistical selection of models of nucleotide substitution online. Nucleic Acids Res. 2006;34:W700–W703. doi: 10.1093/nar/gkl042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 34.Wiens JJ. Missing data and the design of phylogenetic analyses. J. Biomed. Inform. 2006;39:34–42. doi: 10.1016/j.jbi.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fourment M, Gibbs MJ. PATRISTIC: a program for calculating patristic distances and graphically comparing the components of genetic change. BMC Evol. Biol. 2006;6:1. doi: 10.1186/1471-2148-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helgason E, Caugant DA, Lecadet MM, Chen Y, Mahillon J, Lovgren A, Hegna I, Kvaloy K, Kolsto AB. Genetic diversity of Bacillus cereus/B. thuringiensis isolates from natural sources. Curr. Microbiol. 1998;37:80–87. doi: 10.1007/s002849900343. [DOI] [PubMed] [Google Scholar]
- 40.Helgason E, Caugant DA, Olsen I, Kolstø AB. Genetic structure of population of Bacillus cereus and B. thuringiensis isolates associated with periodontitis and other human infections. J. Clin. Microbiol. 2000;38:1615–1622. doi: 10.1128/jcm.38.4.1615-1622.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilas-Boas G, Sanchis V, Lereclus D, Lemos MV, Bourguet D. Genetic differentiation between sympatric populations of Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 2002;68:1414–1424. doi: 10.1128/AEM.68.3.1414-1424.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill KK, Ticknor LO, Okinaka RT, Asay M, Blair H, Bliss KA, Laker M, Pardington PE, Richardson AP, et al. Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl. Environ. Microbiol. 2004;70:1068–1080. doi: 10.1128/AEM.70.2.1068-1080.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson PJ, Hill KK, Laker MT, Ticknor LO, Keim P. Genetic comparison of Bacillus anthracis and its close relatives using amplified fragment length polymorphism and polymerase chain reaction analysis. J. Appl. Microbiol. 1999;87:263–269. doi: 10.1046/j.1365-2672.1999.00884.x. [DOI] [PubMed] [Google Scholar]
- 44.Radnedge L, Agron PG, Hill KK, Jackson PJ, Ticknor LO, Keim P, Andersen GL. Genome differences that distinguish Bacillus anthracis from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 2003;69:2755–2764. doi: 10.1128/AEM.69.5.2755-2764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ticknor LO, Kolsto AB, Hill KK, Keim P, Laker MT, Tonks M, Jackson PJ. Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil isolates. Appl. Environ. Microbiol. 2001;67:4863–4873. doi: 10.1128/AEM.67.10.4863-4873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klee SR, Ozel M, Appel B, Boesch C, Ellerbrok H, Jacob D, Holland G, Leendertz FH, Pauli G, et al. Characterization of Bacillus anthracis-like bacteria isolated from wild great apes from Cote d'Ivoire and Cameroon. J. Bacteriol. 2006;188:5333–5344. doi: 10.1128/JB.00303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim K, Seo J, Wheeler K, Park C, Kim D, Park S, Kim W, Chung SI, Leighton T. Rapid genotypic detection of Bacillus anthracis and the Bacillus cereus group by multiplex real-time PCR melting curve analysis. FEMS Immunol. Med. Microbiol. 2005;43:301–310. doi: 10.1016/j.femsim.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 48.Marston CK, Gee JE, Popovic T, Hoffmaster AR. Molecular approaches to identify and differentiate Bacillus anthracis from phenotypically similar Bacillus species isolates. BMC Microbiol. 2006;6:22. doi: 10.1186/1471-2180-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]