Abstract
The protein property prediction and testing database (PPT-DB) is a database housing nearly 30 carefully curated databases, each of which contains commonly predicted protein property information. These properties include both structural (i.e. secondary structure, contact order, disulfide pairing) and dynamic (i.e. order parameters, B-factors, folding rates) features that have been measured, derived or tabulated from a variety of sources. PPT-DB is designed to serve two purposes. First it is intended to serve as a centralized, up-to-date, freely downloadable and easily queried repository of predictable or ‘derived’ protein property data. In this role, PPT-DB can serve as a one-stop, fully standardized repository for developers to obtain the required training, testing and validation data needed for almost any kind of protein property prediction program they may wish to create. The second role that PPT-DB can play is as a tool for homology-based protein property prediction. Users may query PPT-DB with a sequence of interest and have a specific property predicted using a sequence similarity search against PPT-DB's extensive collection of proteins with known properties. PPT-DB exploits the well-known fact that protein structure and dynamic properties are highly conserved between homologous proteins. Predictions derived from PPT-DB's similarity searches are typically 85–95% correct (for categorical predictions, such as secondary structure) or exhibit correlations of >0.80 (for numeric predictions, such as accessible surface area). This performance is 10–20% better than what is typically obtained from standard ‘ab initio’ predictions. PPT-DB, its prediction utilities and all of its contents are available at http://www.pptdb.ca
INTRODUCTION
Proteins are complex polymers that often defy simple numeric or symbolic descriptions. Their sequences are too long to memorize, their structures are too complex to draw, their motions are too convoluted to animate and their folding processes are too hard to explain. To deal with these ‘fine grain’ complexities we must often resort to describing proteins in terms of their ‘coarse grain’ physical or chemical properties. These coarse-grain properties include such features as radius of gyration, molecular weight, isoelectric point, hydrophobicity, secondary structure, contact order (1), order parameters (2) and folding rates (1,3). In many cases, these physico-chemical properties can be accurately calculated or predicted directly from the protein sequence or the protein's 3D structure (4,4–7). Some properties, such as radius of gyration, molecular weight and isoelectric point can be easily calculated using simple formulas or tables (6), while other properties, such as secondary structure, order parameters and disulfide connectivity are non-trivial to predict or calculate (7,2,5).
The challenges faced in accurately predicting or calculating ‘non-trivial’ protein properties has attracted the interest of many protein chemists, structural biologists and computational biologists for a very long time. Indeed, protein property prediction is one of the oldest disciplines in bioinformatics, with secondary structure prediction being perhaps the earliest (8) and most frequently attempted kind of protein property prediction. Since the 1960s many other kinds of protein properties and property prediction methods have emerged, including methods to predict or identify beta turns (9), membrane helices (10), transmembrane barrel proteins (11), signal peptides (12), disulfide pairings (7) edge or central beta strands (13), beta hairpins (14), coiled-coils (15), accessible surface area (16) and flexibility (17)—to name just a few.
Key to the development of all of these property prediction methods has been the creation or compilation of databases that contain the properties of interest that are to be predicted. Historically these databases served as the raw material from which to derive statistical or heuristic rules about certain protein or amino acid properties (5,18). More recently these databases have served as the testing, training and validation sets for more advanced machine-learning methods (such as neural nets, hidden Markov models and support vector machines) aimed at improving the accuracy of older protein property prediction methods (9–17). In almost all cases, the accuracy of the prediction method is directly dependent on the size, completeness and accuracy of the testing/training database. In other words, protein property databases are absolutely critical to the advancement of protein property prediction.
Unfortunately, the importance of these databases is somewhat underplayed in the bioinformatics community. With the exception of a small number of database resources such as EVA (19), TMH-Benchmark (20), SCRATCH (7) and SPdb (21), very few protein property databases are publicly available or routinely updated. In many cases, protein property databases that were painstakingly assembled by a graduate student or post-doctoral fellow to train their particular predictor are not (or no longer) publicly available. In other cases, if the database is available, it is so woefully out of date or its format is so obscure that it is often more efficient to regenerate a new database from scratch. Still in other cases, the precise origin, method of data generation or the quality of the data is too uncertain to allow the database to be used. Even if a high quality, continuously updated protein property database is available, it is often difficult to locate or access such a resource as there is no common repository for these kinds of databases. As a result most labs that wish to develop, refine or improve upon a given protein property prediction method must resort to ‘re-inventing the wheel’ and generate their own database in their own format. This seems both inefficient and unproductive.
While limited access to high-quality protein property databases is certainly a concern for many data miners and software developers, this limited access also has negative consequence for a large community of users (i.e. scientists wanting predictions). What is not widely appreciated is the fact that protein property databases can also be used as property predictors on their own, especially through the use of homology-based property prediction. This simple method of property prediction is based on the well-known fact that both protein structure and protein dynamic properties are highly conserved between homologous proteins (22). In homology-based property prediction the sequence of interest is aligned against a database of sequences with known properties, features or coordinates. The properties for the highest scoring homolog(s) are then mapped to the query sequence to create a ‘prediction’. Certainly the success of homology or comparative modeling has clearly shown how coordinate mapping from the PDB can be exploited to accurately predict 3D structures for a large number of query proteins. Similar success has been achieved in chemical shift prediction and torsion angle prediction (23,24). More recently, the use of sequence alignments against large databases of proteins of known secondary structure has been shown to improve the quality of secondary structure prediction by a substantial margin (22). Similar improvements in many other property predictions are also possible (vide infra). Obviously these kinds of homology-based predictions are limited to query proteins that exhibit some degree of sequence identity to proteins in the database(s). However, with the size of these protein property databases getting so large, the probability of finding a match is often >60% (22).
Given the importance of protein property predictions and the critical need for high-quality protein property databases, we decided to create an open-access, continuously updated, comprehensive protein property database called the protein Property Prediction and Testing Database (PPT-DB). The intent of this database is to facilitate software development in protein property prediction and to facilitate property prediction by homology. The PPT-DB currently contains nearly 30 carefully curated databases, each of which contains ‘non-trivial’ protein property information. These properties include both structural and dynamic features that have been measured, derived or tabulated from a variety of sources (Table 1). The PPT-DB is designed to serve two purposes. First, it is intended to serve as a centralized, up-to-date, freely downloadable and easily queried repository of predictable or ‘derived’ protein property data. In this role, PPT-DB can serve as a one-stop, standardized (i.e. uniformly formatted and carefully validated) repository for software developers to obtain the required training, testing and validation data needed for almost any kind of protein property prediction program they may wish to create. The second role that PPT-DB can play is as a tool for homology-based protein property prediction. Users may query PPT-DB with a sequence of interest and have a specific property predicted using a sequence similarity search against PPT-DB's collection of proteins with known properties. In many cases these homology-derived property predictions are substantially better than those that would be obtained using conventional or ab initio predictors. A more detailed description of the PPT-DB along with all of its contents and capabilities follows.
Table 1.
Summary of the content and description of different PPT-DB databases
| Database | Description | Database | Description |
|---|---|---|---|
| 2° Structure (cytoplasmic) 15 002 sequences | 3-state 2° structure assignments obtained by VADAR (25) for non-membrane proteins | Signal Peptide (Eukaryotic, Gram+, Gram−) 23 067 sequences | 2-state signal peptide assignments obtained from SwissProt comment fields — grouped via organism type |
| EVA 2° Structure Test Set 7117 sequences | 3-state 2° structure assignments via VADAR (25) for EVA's sequence-unique proteins (19) | Accessible Surface Area (integerized) 14 871 sequences | Residue-specific accessible surface area obtained via VADAR (25) and scaled to values between 0 and 9 |
| 2° Structure (membrane helix) 254 sequences | 2-state 2° structure assignments obtained via VADAR (25) for helical membrane proteins | Accessible Surface Area (%) 14871 sequences | Residue-specific accessible surface area obtained via VADAR (25) and converted to percentage values |
| TMH Benchmark Test Set 2247 sequences | 2-state 2° structure assignments for transmembrane helices from TMH Benchmark (20) | B-factor (integerized) 10 332 sequences | Residue-specific B-factors obtained directly from PDB files of X-ray structures and scaled to values from 0 to 9 |
| 2° Structure (membrane barrel) 41 sequences | 2-state 2° structure assignments obtained by VADAR (25) for trans-membrane barrel proteins | B-factor 10 332 sequences | Residue-specific B-factors obtained directly from PDB (26) files of non-membrane X-ray structures |
| % 2° Structure (cytoplasmic) 15 002 sequences | 3-state 2° structure content obtained by VADAR (25) for non-membrane proteins | RMSF (integerized) 2134 sequences | Scaled (0–9) residue-specific RMSF values determined from NMR structures via SuperPose (29) |
| Beta Turns 14 571 sequences | 5-state beta-turn assignments obtained via VADAR (25) for non-membrane proteins | RMSF 2134 sequences | Residue-specific root mean square fluctuation (RMSF) determined from NMR structures via SuperPose (29) |
| Coiled-coil 824 sequences | 2-state, positional assignments for coiled coil regions from the Paircoil2 training set (15) | Order Parameter (integerized) 9800 sequences | Scaled (0–9) residue-specific order parameter (model free) determined using Contact Model method (2) |
| Edge/Central Beta Strands 13 255 sequences | 2-state beta strand type assignments obtained by VADAR (25) and pattern recognition programs | Order Parameter 9800 sequences | Residue-specific order parameter (model free) determined using Contact Model method (2) |
| Beta Hairpins 8600 sequences | 2-state beta hairpin assignments obtained by VADAR (25) and pattern recognition programs | Contact Order 14 769 sequences | Contact order calculated using method of Plaxco et al. (1) directly from PDB coordinates |
| Disulfide Bonds 2785 sequences | Disulfide bond pairings obtained by VADAR (25) and PDB comment fields | Folding Rate 83 sequences | Experimentally measured folding rates (ln[kf]) obtained from multiple sources (1,3) |
| SPdb (Eukaryotic, Gram+, Gram−) 2590 sequences | Experimentally verified 2-state signal peptide assignments obtained from the SPdb (21) grouped via organism type | 3D Folding Decoys 52 sequences | PDB coordinates for misfolded or improperly folded proteins generated via different 3D prediction tools |
DATABASE DESCRIPTION
PPT-DB is actually a database of databases. As seen in Table 1, PPT-DB consists of 29 smaller sequence databases, each of which contains between 41 and 23 067 Sequences. Altogether the most recent version of the PPT database consists of 234 787 sequences (of which 40 254 are unique), occupying a total of 245 Mb of disk space. So far as we are aware, PPT-DB is the largest and most complete collection of protein property data that has ever been assembled. The protein properties covered in the current release of PPT-DB fall into two broad categories: (i) structural properties and (ii) dynamic properties. The structural properties consist of features that define some aspect of the secondary or tertiary structure of a protein such as secondary structure content, helix location, beta strand location, random coil location, beta turn location, coil–coil location, disulfide bond patterns, signal peptides, contact order, etc. The dynamic properties consist of features that define some aspect of the motion, flexibility or rate processes for a protein, such as root-mean square fluctuation (RMSF), B-factors, folding rate (ln[k]) or order parameters. Some of these properties are global, meaning that one or two numbers describe the property for the entire protein (such as folding rate or contact order). Other properties are local or residue-specific, meaning that individual residues are assigned a value or property (such as secondary structure, B-factors or disulfide pairings).
Every protein property database in PPT-DB, with the exception of the folding decoy database, consists of sequences in a FASTA-like format. The folding decoy database is unique among PPT-DB's property databases as it actually consists of coordinate data rather than pure sequence data. Among the 28 pure sequence databases in the PPT-DB, each sequence is annotated with the name of the sequence, the SwissProt accession number (if it exists) and a PDB accession number (if it exists). Global protein properties (folding rate or contact order) are always displayed on the FASTA header line, while local properties are displayed on a line immediately below the sequence, under the residue(s) with that property (see Figure 1 for examples). For certain properties (B-factors, RMSF, order parameters) the data are stored and displayed in two different formats—horizontally and vertically. The horizontal (FASTA) format requires that the multi-digit numbers be scaled to a single digit integer value between 0 and 9 so that they can be displayed directly underneath the single-character sequence data. The vertical format displays both the sequence and the actual (i.e. multi-digit, real) numbers for the associated property in vertical columns. Additional details about the format, content, scaling and labeling conventions in each database are given in the ‘Database Details’ link located at the top of each database query page.
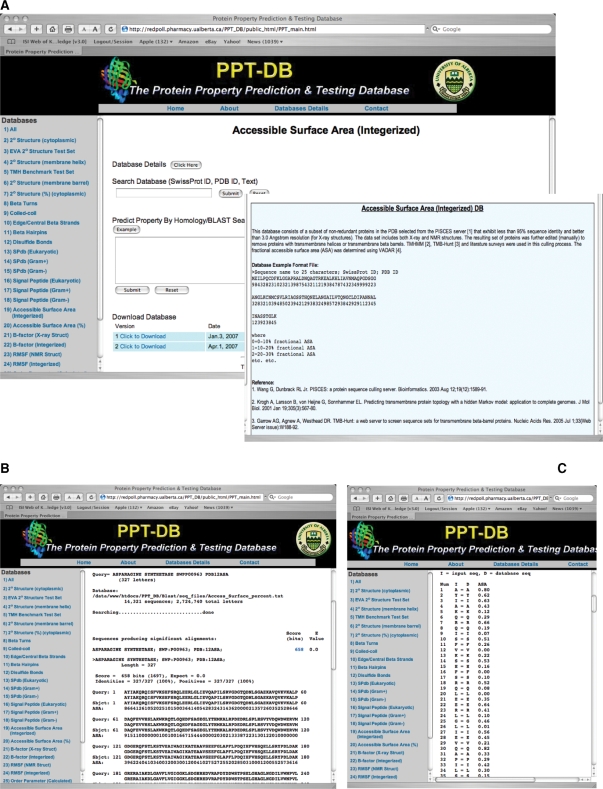
Figure 1.
A screenshot montage showing different windows from the PPT-DB server. (A) An example of a typical sub-database query page, with an example of the content found the ‘Database Details’. (B) and (C) illustrate the two kinds of BLAST search output, with (B) showing the standard horizontal or FASTA format and (C) showing the vertical or column format for displaying residue-specific values/properties that are multi-digit numbers.
As mentioned earlier, the PPT-DB is a dual-purpose resource, serving as either a fully downloadable database for software developers and data miners or as a general property prediction service for protein chemists and structural biologists. Access to both components of PPT-DB is through a relatively simple web interface (Figure 1). As seen in this figure, the left margin of the PPT-DB home page consists of a hyperlinked list of all of its component databases. Clicking on any one of these sub-database hyperlinks generates a database-specific query page (Figure 1A). At the top of each database query page is the name of the protein property database followed by a ‘Database Details’ button. Clicking on this button provides detailed information on the database format and how the database was constructed (Figure 1A). Below this (with the exception of the folding decoy database) is a text search box. Users may search their selected database using the protein name (or part thereof), the SwissProt ID (or part thereof), PDB ID (or part thereof) or any other text within the sequence header. This text search will rapidly generate a 3-column hyperlinked table showing the name of the protein, the SwissProt ID and the PDB ID. Clicking on the protein name will display that proteins sequence along with its PPT-DB property annotations. Clicking on the SwissProt ID will open the corresponding SwissProt entry for that protein while clicking on the PDB ID will open the corresponding PDB web page for that protein.
The PPT-DB is also searchable via sequence queries using a local version of BLAST. However, the main purpose of PPT-DB's BLAST search is not necessarily to locate a given protein in the database, but rather to predict the properties of a query (i.e. an unknown or uncharacterized) protein through a technique known as homology-based property prediction or homology-based property mapping (22–24). In other words, the BLAST sequence search is part of PPT-DB's general protein property prediction service. This service is primarily intended for protein chemists, molecular biologists and structural biologists. Details concerning the performance of these property predictions are described in the ‘Database Details’ link at the top of each database query page, as well as in the section entitled ‘Protein Property Prediction using PPT-DB’ presented later in this manuscript. PPT-DB's BLAST search accepts both FASTA and ‘raw’ sequence data as input and uses a default E (expect) value of 10−5 as a cutoff for selecting sequence matches. Each BLAST query in the PPT-DB has an ‘Example’ button which uploads a sample sequence, allowing new users to test PPT-DB's search utilities and investigate the output format for each kind of database query. Figure 1B and C illustrate the type of output generated by PPT-DB's BLAST search, showing examples of both the horizontal and vertical output formats. The PPT-DB also supports BLAST queries against all of PPT-DB's protein properties through the ‘All Properties’ database located at the top of the database list. This particular feature allows users to ‘predict’ all PPT-DB properties (24 of 28) that can be displayed in a FASTA (horizontal) format.
At the bottom of each database query page is the database download page. This is a hyperlinked table providing information about the version number, the release date, the size and number of sequences in each version of a given PPT database. Pressing on the ‘Click to Download’ link allows users to retrieve or install a local copy of each PPT sub-database on their own computer. The structure and annotation details for each version of every PPT-DB sub-database are contained in a header at the top of each database file. All of PPT-DB's databases are stored as simple, uncompressed text files. The ‘Download’ section of the PPT-DB is primarily intended to support the activities of software developers interested in training, testing and validating their property prediction software or data miners interested in extracting statistics, trends or heuristics about certain protein properties. As described in the following section, every effort is made to ensure that these downloadable databases are as current, complete and correct as possible.
DATABASE PREPARATION, QUALITY ASSURANCE AND CURATION
Table 1 briefly describes the sources, protocols or programs used to generate most of the databases in PPT-DB. Additional details concerning the preparation and maintenance of each database are provided in the ‘Database Details’ link located in PPT-DB's menu bar or via the ‘Database Details’ button located within each PPT-DB sub-database. As seen from Table 1, many of the databases created for PPT-DB were developed internally using a program called VADAR (25) to help derive or extract the data from existing PDB files. Other PPT databases were assembled from information contained explicitly in SwissProt (26) or the PDB (27). In these cases, database-specific programs were written to extract data from SwissProt headers or PDB comment fields and to map this information on to the sequence extracted from the corresponding database. Essentially, all databases that were primarily derived from the PDB (or subsequently by VADAR) were processed by the PDB culling/filtering service called PISCES (28). Structures were selected using a 95% identity sequence-redundancy cutoff and a requirement for better than 3 Å resolution (for X-ray structures). These files were further filtered to remove electron microscopy models and protein chains having fewer than 30 residues.
While the vast majority of the databases in PPT-DB were derived using automatic methods, some databases (such as the transmembrane helix, the transmembrane beta barrel database, the folding rate and folding decoy database) were assembled manually. Other databases, such as the beta hairpin and the beta edge/central strand were created using specialized programs that re-interpreted standard VADAR output. Still other databases such as EVA (19), SPdb (21), the Coiled–Coil/Paircoil2 database (15) and TMH-Benchmark (20) were obtained from external sources but were re-formatted and manually edited or upgraded to make them compatible with the PPT-DB annotation standards.
Each database and each update to a database is numbered and dated allowing a well-defined audit trail to be assembled. This is intended to allow external software developers and external data miners the opportunity to share and compare testing/training data. With the exception of the signal peptide databases, almost all sequences in the PPT-DB were derived from the original PDB sequence file. As a consequence, the sequence for the corresponding SwissProt entry may sometimes differ from the sequence listed in either the PPT-DB or the PDB. All databases, with the exception of the folding decoys database and the folding rate database, have automated or semi-automated scripts to facilitate updating. Depending on their size and ease of curation, PPT-DB databases are updated as frequently as once per month (i.e. secondary structure databases) or as infrequently as once per year (i.e. the folding rate database).
In preparing and updating the PPT-DB, every effort has been made to ensure that each database is as complete, correct and current as possible. Certainly many of the programs used in the data generation process, such as VADAR (25) and SuperPose (29), have had more than a decade of testing and are considered very robust. Nevertheless, a ‘PPT-DB sanity checker’ has been written to ensure that impossible numeric values or disallowed characters are flagged in any existing database and any subsequent updates. These problem entries are then manually assessed and manually corrected as required. As a further quality control measure, spot checks are routinely performed on many entries by senior members of the curation group, including two PhD-level biochemists.
PROPERTY PREDICTION USING PPT-DB
One of the most useful and important applications of PPT-DB lies in its ability to help predict protein properties through homology-based property mapping. Just as sequence searches through GenBank and SwissProt allow evolutionary relationships or functional annotations to be made for newly sequenced proteins, so too it is possible to use sequence searches through PPT-DB to accurately predict both structural and dynamic properties of proteins. Previous studies have shown that homology-based property prediction can significantly outperform the best ab initio (neural net, SVM or HMM) prediction methods—if the query is sufficiently similar to a protein in the database (22–24). These homology-based predictions are also very fast (<1 s) compared to most other advanced property prediction methods (most of which take minutes). However, one obvious limitation of homology-based property prediction is that it only works if some level of sequence homology exists. Surprisingly, this requirement is not as onerous or as infrequent as one might think.
To evaluate the performance of each of the PPT-DB's property predictors, we used a limited 10-fold cross-validation assessment. Specifically, we randomly selected 10 sets of 100 proteins (or fewer if the database had <1000 sequences) from each PPT database and used these as queries for the corresponding PPT-DB BLAST search using an expect value cutoff of 10−5. After the exact match was excluded, the second highest scoring hit (if such a hit was found) was used to predict the property of the query protein. The prediction was then scored using standard Q2 or Q3 methods (i.e. % correct) for categorical predictions, such as secondary structure, or correlation coefficients for numeric predictions, such as accessible surface area. For global properties, such as folding rate, contact order or secondary structure content, the prediction is only accepted if the sequence length of the matching protein is 100 ± 20% of the query protein's length. A similar rule is also used for accessible surface area predictions. Results for each of these 10 prediction sets were tabulated, both in terms of performance (average and standard deviation) and overall coverage. Results for individual prediction sets were also plotted using scatter plots (performance versus sequence identity) along with the proportion of queries that exhibited significant hits (i.e. the coverage). All of these scatter plots and performance statistics are available by clicking the ‘Database Details’ button for each PPT sub-database. Figure 2 illustrates the results obtained for secondary structure prediction via PPT-DB. This is a fairly typical result with the performance generally dropping as the sequence identity drops below 35–40%. As noted in this figure, PPT-DB achieves an average Q3 of 89.1%. This compares quite favorably to Q3's of 75% reported for most conventional secondary structure prediction methods (5, 22). Similar kinds of results are obtained for many other protein properties. For instance, PPT-DB's accessible surface area (ASA) prediction achieves a correlation coefficient of 84.5%, which is between 8 and 15% better than the best methods for ASA (3-state only) prediction (16). Likewise PPT-DB obtains a Q2 for beta hairpin prediction of 93.0%, while the best ab initio method attains a Q2 of 77% (14). PPT-DB also performs quite well in identifying edge/central beta strands with a predictive accuracy of 90.1% compared to only 55% for the best ab initio method (13). Space limitations prevent a detailed comparison between all of PPT-DB's property predictions and those reported in the literature. However, interested readers can view the performance statistics by clicking the ‘Database Details’ button for each PPT sub-database. As seen from the tables and scatter plots in these ‘Database Details’ pages, predictions derived from PPT-DB's similarity searches are typically 85–95% correct (for categorical or global property predictions) or exhibit correlations of >0.80 (for numeric predictions). This performance is typically 10–20% better than what is obtained from the best ‘ab initio’ predictions. Our data also suggests that reliable PPT-DB predictions are possible for ∼75% of all query sequences, as long as the database contains >10 000 sequences. Certainly as PPT-DB's databases increase in size, the property prediction performance and level of coverage would be expected to increase as well.
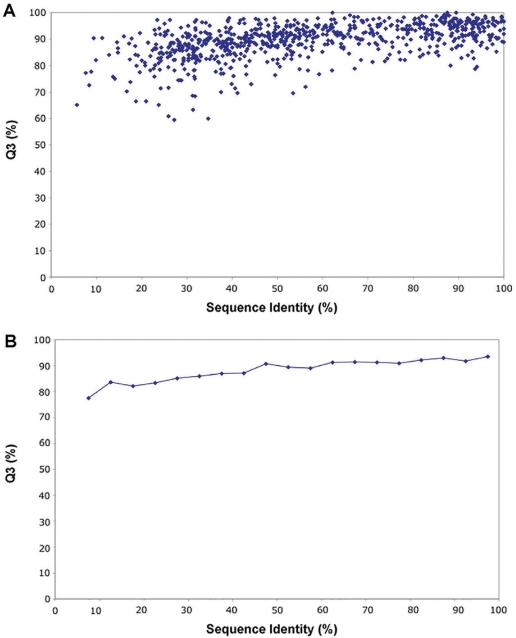
Figure 2.
(A) A scatter plot showing of the predictive performance (Q3 versus% sequence identity) for secondary structure prediction for 1000 random query sequences that were submitted to PPT-DB's secondary structure database. (B) A moving average of the same data shown in A (using 5% sequence identity intervals). Using 10-fold cross-validation (10 sets of 100 random query proteins), the average coverage was 77.1 ± 9.3% and the average Q3 for all secondary structure predictions was 89.1 ± 1.3%.
CONCLUSION
In summary, the PPT-DB is a comprehensive, open-access, continuously updated, protein property database. It was developed to fill a database void in the field of protein property prediction, which currently lacks both a uniformly formatted and a centralized repository of known or predicted protein properties. The PPT-DB was also designed to appeal to two very different audiences: (i) programmers and (ii) biologists. Software developers should find it helpful in developing, testing, comparing and improving their own protein property prediction programs while structural biologists and protein chemists should find it useful as a fast and accurate tool to help predict a wide range of important protein properties. While the protein properties covered by the PPT-DB are comprehensive, they certainly are not complete. Over the coming year we are hoping to add more property databases, including protein ‘site’ modification databases (glycosylation, sulfation, phosphorylation sites), protein solubility databases, thermostability databases, subcellular location databases and amyloid propensity databases. Likewise we are certainly open to suggestions for new kinds of databases or new database formats. Submissions from external sources of new protein property databases to the PPT-DB (if appropriately formatted, described and validated) are welcomed. Overall it is hoped that the PPT-DB will help improve the quality and reliability of protein property predictions as well as the frequency with which these kinds of predictions are made or reported in the literature.
ACKNOWLEDGEMENTS
The authors wish to thank the Alberta Prion Research Institute (APRI), PrioNet (a National Centre of Excellence), NSERC and Genome Alberta (in association with Genome Canada) for financial support. Funding to pay the Open Access publication charges for this article was provided by Genome Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J. Mol. Biol. 1998;77:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 2.Zhang F, Bruschweiler R. Contact model for the prediction of NMR N-H order parameters in globular proteins. J. Am. Chem. Soc. 2002;124:12654–12655. doi: 10.1021/ja027847a. [DOI] [PubMed] [Google Scholar]
- 3.Fulton KF, Bate MA, Faux NG, Mahmood K, Betts C, Buckle AM. Protein Folding Database (PFD 2.0): an online environment for the International Foldeomics Consortium. Nucleic Acids Res. 2007;35(Database issue):D304–D307. doi: 10.1093/nar/gkl1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wishart DS. Tools for protein technologies. In: Rehm HJ, Reed G, Puhler A, Stadler P, editors. Genomics and Bioinformatics Biotechnology. 2nd. Weinheim: Wiley-VCH; 2001. pp. 326–342. [Google Scholar]
- 5.Fasman GD. Prediction of Protein Structure and the Principles of Protein Conformation. New York, NY: Plenum Press; 1989. [Google Scholar]
- 6.Richards FM. Areas, volumes, packing and protein structure. Annu. Rev. Biophys. Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 7.Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33(Web Server issue):W72–W76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guzzo AV. The influence of amino acid sequence on protein structure. Biophys. J. 1965;5:809–822. doi: 10.1016/S0006-3495(65)86753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, Yoon S, Welsh WJ. Improved method for predicting beta-turn using support vector machine. Bioinformatics. 2005;21:2370–2374. doi: 10.1093/bioinformatics/bti358. [DOI] [PubMed] [Google Scholar]
- 10.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 11.Garrow AG, Agnew A, Westhead DR. TMB-Hunt: a web server to screen sequence sets for transmembrane beta-barrel proteins. Nucleic Acids Res. 2005;33(Web Server issue):W188–W192. doi: 10.1093/nar/gki384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 13.Siepen JA, Radford SE, Westhead DR. Beta edge strands in protein structure prediction and aggregation. Protein Sci. 2003;12:2348–2359. doi: 10.1110/ps.03234503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar M, Bhasin M, Natt NK, Raghava GP. BhairPred: prediction of beta-hairpins in a protein from multiple alignment information using ANN and SVM techniques. Nucleic Acids Res. 2005;33(Web Server issue):W154–W159. doi: 10.1093/nar/gki588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- 16.Dor O, Zhou Y. Real-SPINE: an integrated system of neural networks for real-value prediction of protein structural properties. Proteins. 2007;68:76–81. doi: 10.1002/prot.21408. [DOI] [PubMed] [Google Scholar]
- 17.Schlessinger A, Rost B. Protein flexibility and rigidity predicted from sequence. Proteins. 2005;61:115–126. doi: 10.1002/prot.20587. [DOI] [PubMed] [Google Scholar]
- 18.Wilmot CM, Thornton JM. Analysis and prediction of the different types of beta-turn in proteins. J. Mol. Biol. 1988;203:221–232. doi: 10.1016/0022-2836(88)90103-9. [DOI] [PubMed] [Google Scholar]
- 19.Rost B, Eyrich VA. EVA: large-scale analysis of secondary structure prediction. Proteins. 2001;(Suppl. 5):192–199. doi: 10.1002/prot.10051. [DOI] [PubMed] [Google Scholar]
- 20.Kernytsky A, Rost B. Static benchmarking of membrane helix predictions. Nucleic Acids Res. 2003;31:3642–3644. doi: 10.1093/nar/gkg532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choo KH, Tan TW, Ranganathan S. SPdb – a signal peptide database. BMC Bioinformatics. 2005;6:249. doi: 10.1186/1471-2105-6-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montgomerie S, Sundararaj S, Gallin WJ, Wishart DS. Improving the accuracy of protein secondary structure prediction using structural alignment. BMC Bioinformatics. 2006;7:301. doi: 10.1186/1471-2105-7-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wishart DS, Watson MS, Boyko RF, Sykes BD. Automated 1H and 13C chemical shift prediction using the BioMagResBank. J. Biomol. NMR. 1997;10:329–336. doi: 10.1023/a:1018373822088. [DOI] [PubMed] [Google Scholar]
- 24.Berjanskii MV, Neal S, Wishart DS. PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 2006;34(Web Server issue):W63–69. doi: 10.1093/nar/gkl341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003;31:3316–3319. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35(Database issue):D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Donovan C, Martin MJ, Gattiker A, Gasteiger E, Bairoch A, Apweiler R. High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Brief Bioinform. 2002;3:275–284. doi: 10.1093/bib/3.3.275. [DOI] [PubMed] [Google Scholar]
- 28.Wang G, Dunbrack R.L., Jr. PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 29.Maiti R, Van Domselaar GH, Zhang H, Wishart DS. SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 2004;32(Web Server issue):W590–W594. doi: 10.1093/nar/gkh477. [DOI] [PMC free article] [PubMed] [Google Scholar]