Abstract
Gallus GBrowse (http://birdbase.net/cgi-bin/gbrowse/gallus/) provides online access to genomic and other information about the chicken, Gallus gallus. The information provided by this resource includes predicted genes and Gene Ontology (GO) terms, links to Gallus In Situ Hybridization Analysis (GEISHA), Unigene and Reactome, the genomic positions of chicken genetic markers, SNPs and microarray probes, and mappings from turkey, condor and zebra finch DNA and EST sequences to the chicken genome. We also provide a BLAT server (http://birdbase.net/cgi-bin/webBlat) for matching user-provided sequences to the chicken genome. These tools make the Gallus GBrowse server a valuable resource for researchers seeking genomic information regarding the chicken and other avian species.
INTRODUCTION
The chicken (Gallus gallus) has played important roles in both scientific research and the general health and welfare of humans. For example, in the field of developmental biology, the chicken embryo model has provided insight into many developmental processes including cell migration (1–3), limb development (4,5) and eye formation (6–8). The discovery of avian oncogenic viruses helped highlight the importance of specific genes in tumorigenesis and the chicken continues to be a popular model system for cancer and other diseases (9–11). As a food source, the chicken was domesticated in Asia ∼7000–10 000 years ago and has undergone intensive selection for both egg and meat production over the past 60–70 years. In 2005, the United States (source: USDA National Agricultural Statistics Service) alone produced and consumed 30 billion pounds and exported another 5 billion pounds of chicken meat. In that same year, 90 billion eggs were produced in the United States. Clearly, the chicken plays an important role as both a model organism and as a food resource.
An enormous amount of genomic information and resources are available for the chicken. The genomic sequence of ∼1 billion nucleotides was completed (12) and released in 2004 and then updated in 2006. A total of 3 335 290 SNPS (13) have been deposited in GenBank and over 1000 microsatellite (MS) and other genetic markers have been identified (14,15). At least five microarray platforms are available, and the Gallus In Situ Hybridization Analysis (GEISHA) (16) project is providing detailed descriptions of the embryonic expression pattern of many chicken genes. A centralized, web accessible, chicken database would provide a valuable resource for common access to this data. To begin providing such a resource, we have developed a Generic Model Organism Database (GMOD) (17) Gallus GBrowse site along with a BLAT server for searching the chicken genome. This site provides access to many chicken resources, along with mappings of turkey, condor and zebra finch nucleotide sequences to the chicken genome.
GALLUS GBROWSE DATA
The draft chicken genomic sequence (V2.1), produced by the Genome Sequencing Center at Washington University of St. Louis, was downloaded from the UCSC Genome Browser Gateway. The GMOD GBrowse viewer (17) in combination with a MySQL database management system is used to store, search and display annotation of the chicken genome. The GBrowse web page provides user access and is organized along themes including genes, gene expression platforms, gene expression data, Gene Ontology (GO) and pathways, markers and SNPs and other avian species.
Genes
The gene positions were defined based upon NCBI RefSeq and Ensembl cDNA predictions. These are provided as separate tracks in the GBrowse. In addition, predicted non-coding RNA genes and exon/intron positions are provided based on Ensembl predictions.
Gene expression platforms
These allow visualizing the positions of probes from five array platforms in the context of the chicken genome. Probe sequences for the Delmar (18), Avian Macrophage (19,20), Chicken 13K (21) and the Chicken Oligo microarray (http://www.grl.steelecenter.arizona.edu/products.asp) were aligned with the chicken genomic sequence using BLAT (22). The probe positions for the Affymetrix Chicken Genome Array were obtained from the NetAffx alignment file provided by Affymetrix.
Gene expression
Currently, two sets of gene expression data are accessed from Gallus GBrowse: GEISHA (16) and Unigene (23). The GEISHA project aims to describe the expression pattern of genes in the chicken embryo between Hamburger and Hamilton stages 1–25. The Unigene information is derived from the Unigene expression profiler, which describes the expression pattern for a gene based on EST analysis.
Gene ontologies and pathways
One set of tracks displays GO (24,25) terms for a given gene. GO terms were obtained from the Gene Ontology Annotation (GOA) Database via the NCBI database gene2go file. Hovering the mouse over the glyph will display the assigned GO term, while clicking on the link will connect to the Amigo term definition.
Reactome (26,27) is a human-centric curated knowledge base of biological pathways and pathways for other species are predicted by gene ortholog relationships. The Gallus GBrowse Reactome glyph links to the gene summary page in the Reactome knowledge base for the corresponding chicken gene. From the Reactome summary page, one can then access all pertinent information regarding the gene, including the reactions, pathways and molecular complexes the gene product participates in, as well as the gene's orthologs in human and other model species.
Markers and SNPS
Markers were obtained from the NCBI UniSTS ftp site, or from a sequence file provided by Dr Martien Groenen (Wageningen University). The genomic locations of these sequences were then determined by BLAT analysis. SNPs were also mapped to the genome by BLAT using the flanking sequence obtained from the NCBI dbSNP database. Because of the high density of SNPs mapped (>3 000 000) to the chicken genome, the SNP track is only visualized at a zoom scale of 250 000 nucleotides or lower. Clicking on an individual SNPs glyph will link to the NCBI cluster report for that SNP.
Other avian species
To help integrate analysis of the chicken with other avian species, genomic and cDNA data from the turkey (28–31), condor and zebra finch (32,33) have been mapped to the chicken genome by BLAT. Turkey DNA and zebra finch DNA sequences were obtained from NCBI along with the condor MS sequences. The condor 454 sequences were derived from fibroblast ESTs determined using the 454 sequencing technology (34).
DNA
This track visualizes the DNA sequence of the current region. The nucleotide sequence is only presented at a zoom of 100 base pairs. At higher zoom levels, the %GC content is displayed.
QUERY TOOLS
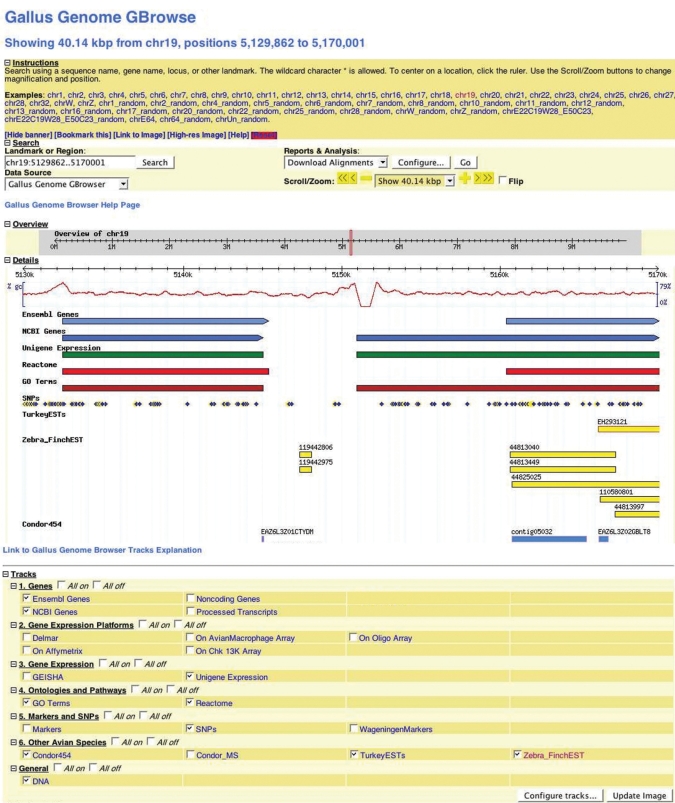
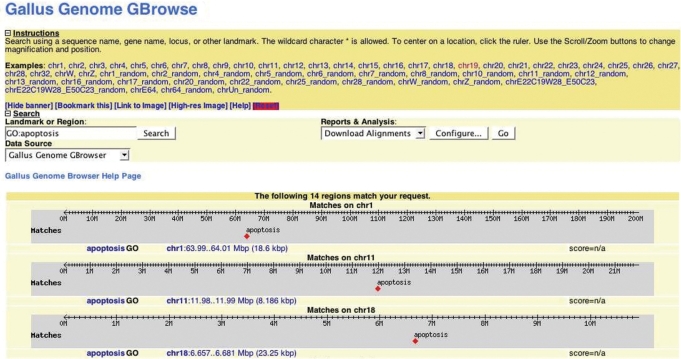
The Gallus GBrowse web page provides an integrated query interface. Specific chromosomal regions of 10 megabases or less can be accessed with known nucleotide coordinates using the Landmark or Region search box (Figure 1). This same search box can be used to locate specific information stored in the GBrowse database. For example, one can search for all genes annotated with the GO term ‘apoptosis’ by inserting ‘GO:apoptosis’ in the Landmark or Region box (Figure 2). This yields a total of 14 genes that have been annotated with ‘apoptosis’ in the chicken genome. A complete listing of all query prefix terms (such as GO) is provided in the Gallus GBrowse help pages.
Figure 1.
Gallus GBrowse. A portion of chicken chromosome 19 (nucleotides 5129862–5170001) shown with glyphs depicting predicted genes (chicken), links to Unigene, Reactome and Gene Ontology annotation, SNPs and the location of turkey, zebra finch and condor ESTs that have been mapped to the chicken genome.
Figure 2.
Searching Gallus GBrowse for specific entries. The search term GO:apoptosis was entered in the Landmark or Region box followed by pushing the Search button. A total of 14 entries were found, only three are presented here. Using the mouse to click on the chromosome link will open the browser to that location.
One of the more challenging aspects of using many genomic databases is searching based on a gene name. As a convention, Gallus GBrowse uses chicken gene names assigned by NCBI and entering a search in the syntax ‘NCBI:gene name’ will typically recover the desired information. Another approach can be to use a homologous nucleotide or protein sequence and the BLAT server (below) to identify the chromosomal location of the gene of interest.
A BLAT server is provided (http://birdbase.net/cgi-bin/webBlat) to allow searching the chicken genome with either nucleotide or protein sequences. Two databases are provided, one containing the nucleotide sequence (Chicken Genome untranslated) and the other containing the chicken genome translated in all reading frames (Chicken Genome translated). To successfully execute a BLAT search, the appropriate database must be selected for nucleotide (untranslated) or protein (translated) input sequence. Results from the BLAT analysis are returned as two web links, one showing the alignment of the query sequence with the matched chicken genomic sequence, and the second displaying the Gallus GBrowse viewer focused on the region of the aligned query sequence.
FUTURE DIRECTIONS
The Gallus GBrowse will be updated as new relevant information becomes available. One near term objective is to incorporate the position of repetitive sequence elements into the GBrowse database. An additional goal is incorporating both microarray and high-throughput EST sequencing data to describe gene expression patterns. Initially this will likely to reflect a simple interpretation of whether or not a gene was detected above background and allow users to determine if a given gene is expressed under the experimental conditions of the microarray or sequencing assay. Gallus GBrowse will also be improved by linking genes with the curated ontology efforts of AgBase (35,36). The current GO entries are derived from uncurated, electronic annotation and the AgBase effort should provide a far more reliable and accurate assignment of GO terms. Finally, a long-term goal is to continue incorporating genomic information from other avian species with the adoption of additional GMOD tools and the Chado database schema. We hope to ultimately provide an integrated resource for comparative avian genomics.
ACKNOWLEDGEMENTS
We are particularly grateful to Drs Parker Antin, Shane Burgess, Greg Keane, Fiona McCarthy and Kent Reed for their input and encouragement. This work was supported, in part, by the University of Delaware Avian Biosciences Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dupin E, Ziller C, Le Douarin NM. The avian embryo as a model in developmental studies: chimeras and in vitro clonal analysis. Curr. Top. Dev. Biol. 1998;36:1–35. doi: 10.1016/s0070-2153(08)60493-7. [DOI] [PubMed] [Google Scholar]
- 2.Funk PE, Thompson CB. Current concepts in chicken B cell development. Curr. Top. Microbiol. Immunol. 1996;212:17–28. doi: 10.1007/978-3-642-80057-3_3. [DOI] [PubMed] [Google Scholar]
- 3.Qureshi MA, Heggen CL, Hussain I. Avian macrophage: effector functions in health and disease. Dev. Comp. Immunol. 2000;24:103–119. doi: 10.1016/s0145-305x(99)00067-1. [DOI] [PubMed] [Google Scholar]
- 4.Tickle C. Morphogen gradients in vertebrate limb development. Semin. Cell Dev. Biol. 1999;10:345–351. doi: 10.1006/scdb.1999.0294. [DOI] [PubMed] [Google Scholar]
- 5.Viallet J, Garcia A, Weydert A. Protein phosphatase 2A as a new target for morphogenetic studies in the chick limb. Biochimie. 2003;85:753–762. doi: 10.1016/j.biochi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers PH, Jamrich M. Regulation of vertebrate eye development by Rx genes. Int. J. Dev. Biol. 2004;48:761–770. doi: 10.1387/ijdb.041878tb. [DOI] [PubMed] [Google Scholar]
- 7.Glover JC. The development of vestibulo-ocular circuitry in the chicken embryo. J. Physiol. Paris. 2003;97:17–25. doi: 10.1016/j.jphysparis.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Naito J, Chen Y. Morphological features of chick retinal ganglion cells. Anat. Sci. Int. 2004;79:213–225. doi: 10.1111/j.1447-073x.2004.00084.x. [DOI] [PubMed] [Google Scholar]
- 9.Davison TF. The immunologists’ debt to the chicken. Br. Poult. Sci. 2003;44:6–21. doi: 10.1080/0007166031000085364. [DOI] [PubMed] [Google Scholar]
- 10.Witter RL. Avian tumor viruses: persistent and evolving pathogens. Acta Vet. Hung. 1997;45:251–266. [PubMed] [Google Scholar]
- 11.Fadly AM. Avian retroviruses. Vet. Clin. North Am. Food Anim. Pract. 1997;13:71–85. doi: 10.1016/s0749-0720(15)30365-0. [DOI] [PubMed] [Google Scholar]
- 12.International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 13.Wong GK, Liu B, Wang J, Zhang Y, Yang X, Zhang Z, Meng Q, Zhou J, Li D, et al. A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature. 2004;432:717–722. doi: 10.1038/nature03156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fillon V, Vignoles M, Garrigues A, Pitel F, Morisson M, Crooijmans RP, Groenen MA, Gellin J, Vignal A. The chicken cytogenetic map: an aid to microchromosome identification and avian comparative cytogenetics. Br. Poult. Sci. 2003;44:795–797. doi: 10.1080/00071660410001666844. [DOI] [PubMed] [Google Scholar]
- 15.Groenen MA, Cheng HH, Bumstead N, Benkel BF, Briles WE, Burke T, Burt DW, Crittenden LB, Dodgson J, et al. A consensus linkage map of the chicken genome. Genome Res. 2000;10:137–147. doi: 10.1101/gr.10.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell GW, Yatskievych TA, Antin PB. GEISHA, a whole-mount in situ hybridization gene expression screen in chicken embryos. Dev. Dyn. 2004;229:677–687. doi: 10.1002/dvdy.10503. [DOI] [PubMed] [Google Scholar]
- 17.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cogburn LA, Wang X, Carre W, Rejto L, Aggrey SE, Duclos MJ, Simon J, Porter TE. Functional genomics in chickens: development of integrated-systems microarrays for transcriptional profiling and discovery of regulatory pathways. Comp. Funct. Genom. 2004;5:253–261. doi: 10.1002/cfg.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bliss TW, Dohms JE, Emara MG, Keeler C.L., Jr Gene expression profiling of avian macrophage activation. Vet. Immunol. Immunopathol. 2005;105:289–299. doi: 10.1016/j.vetimm.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Keeler CL, Bliss TW, Lavric M, Maughan MN. A functional genomics approach to the study of avian innate immunity. Cytogenet. Genome Res. 2007;117:139–145. doi: 10.1159/000103174. [DOI] [PubMed] [Google Scholar]
- 21.Burnside J, Neiman P, Tang J, Basom R, Talbot R, Aronszajn M, Burt D, Delrow J. Development of a cDNA array for chicken gene expression analysis. BMC Genomics. 2005;6:13. doi: 10.1186/1471-2164-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:5–12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:322–326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vastrik I, D’Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaves LD, Rowe JA, Reed KM. Survey of a cDNA library from the turkey (Meleagris gallopavo) Genome. 2005;48:12–17. doi: 10.1139/g04-088. [DOI] [PubMed] [Google Scholar]
- 29.Reed KM, Knutson TP, Krueth SB, Sullivan LR, Chaves LD. In silco mapping of ESTs from the turkey (Meleagris gallopavo) Anim. Biotechnol. 2005;16:81–8102. doi: 10.1080/10495390500261470. [DOI] [PubMed] [Google Scholar]
- 30.Dranchak PK, Chaves LD, Rowe JA, Reed KM. Turkey microsatellite loci from an embryonic cDNA library. Poult. Sci. 2003;82:526–531. doi: 10.1093/ps/82.4.526. [DOI] [PubMed] [Google Scholar]
- 31.Smith E, Shi L, Drummond P, Rodriguez L, Hamilton R, Powell E, Nahashon S, Ramlal S, Smith G, et al. Development and characterization of expressed sequence tags for the turkey (Meleagris gallopavo) genome and comparative sequence analysis with other birds. Anim. Genet. 2000;31:62–67. doi: 10.1046/j.1365-2052.2000.00590.x. [DOI] [PubMed] [Google Scholar]
- 32.Wade J, Peabody C, Coussens P, Tempelman RJ, Clayton DF, Liu L, Arnold AP, Agate R. A cDNA microarray from the telencephalon of juvenile male and female zebra finches. J. Neurosci. Methods. 2004;138:199–206. doi: 10.1016/j.jneumeth.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Wang X.-J, Tannenhauser J, Podell S, Mukherjee P, Hertel M, Biane J, Masuda S, Nottebohm F, et al. Genomic resources for songbird research and their use in characterizing gene expression during brain development. Proc. Natl Acad. Sci. USA. 2007;104:6834–6839. doi: 10.1073/pnas.0701619104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Al E. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy FM, Bridges SM, Wang N, Magee GB, Williams WP, Luthe DS, Burgess SC. AgBase: a unified resource for functional analysis in agriculture. Nucleic Acids Res. 2007;35:D599–D603. doi: 10.1093/nar/gkl936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy FM, Wang N, Magee GB, Nanduri B, Lawrence ML, Camon EB, Barrell DG, Hill DP, Dolan ME, et al. AgBase: a functional genomics resource for agriculture. BMC Genomics. 2006;7:229. doi: 10.1186/1471-2164-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]