Abstract
The BioHealthBase Bioinformatics Resource Center (BRC) (http://www.biohealthbase.org) is a public bioinformatics database and analysis resource for the study of specific biodefense and public health pathogens—Influenza virus, Francisella tularensis, Mycobacterium tuberculosis, Microsporidia species and ricin toxin. The BioHealthBase serves as an extensive integrated repository of data imported from public databases, data derived from various computational algorithms and information curated from the scientific literature. The goal of the BioHealthBase is to facilitate the development of therapeutics, diagnostics and vaccines by integrating all available data in the context of host–pathogen interactions, thus allowing researchers to understand the root causes of virulence and pathogenicity. Genome and protein annotations can be viewed either as formatted text or graphically through a genome browser. 3D visualization capabilities allow researchers to view proteins with key structural and functional features highlighted. Influenza virus host–pathogen interactions at the molecular/cellular and systemic levels are represented. Host immune response to influenza infection is conveyed through the display of experimentally determined antibody and T-cell epitopes curated from the scientific literature or as derived from computational predictions. At the molecular/cellular level, the BioHealthBase BRC has developed biological pathway representations relevant to influenza virus host–pathogen interaction in collaboration with the Reactome database (http://www.reactome.org).
INTRODUCTION
Seasonal flu is an acute viral infection generally involving the upper respiratory tract that affects 5–20% of the human population resulting in the death of ∼35 000 people each year in the US. Although mortality rates from flu are typically low (<0.1%) (1), three times during the last century an especially virulent form of the disease emerged, resulting in pandemics. In 1918, the Spanish flu (subtype H1N1) swept across Europe and the United States causing 40–50 million deaths (2). In 1957 and 1968, the Asian flu (H2N2) and Hong Kong flu (H3N2) claimed ∼1 million lives each.
Influenza structure
In order to understand, and ultimately prevent, the emergence of these deadly pandemics it is essential to understand the key characteristics of the etiologic agent and the nature of how it interacts with its hosts at the molecular level. Influenza virus is a member of the Orthomyxoviridae family of segmented negative single-stranded RNA viruses. The genome of Influenza A virus is composed of eight RNA segments, which together encode 11 functional polypeptides (3). Many of the influenza virus proteins contribute to the virus host range. The PA, PB1, PB2 and NP proteins form an RNA polymerase complex responsible for viral RNA replication and transcription. The NP protein also coats the viral RNA genome segments to form the ribonucleoprotein (RNP) core. The HA protein facilitates virion binding to sialic acid glycolipids on the host cell's plasma membrane and also facilitates endosome fusion through an acid-induced conformation change mechanism. The NA protein facilitates virion release through its neuraminidase activity. The NS1 protein plays a critical role in facilitating viral replication by inhibiting the host immune response to viral infection. The remaining proteins M1, M2 and NS2 function as structural proteins while PB1-F2 assists in apoptosis.
Host range
As a species, influenza virus can infect a variety of mammalian and non-mammalian hosts, including wild and domesticated birds, pigs and humans. However, individual viral isolates exhibit more selective host range preferences (4). Host-range specificity appears to be partly dictated by the complementarities between variants of the viral HA proteins and the structure of the sialic acid on the host cell surface (5). More recently, other influenza proteins have also been found to influence host range to varying degrees.
Viral evolution
While influenza virus has developed a variety of mechanisms to dampen the initial immune response to viral infection, the virus is ultimately eliminated through a combination of innate and adaptive immune responses (6). But if protective immunity against influenza is routinely elicited, why are we susceptible to the disease each year, and how does a pandemic strain emerge on occasion? The answers to these questions relate to the nature and evolution of the viral genome, and two phenomena of HA variation—antigenic drift and antigenic shift (7).
As with all other species, influenza evolves through a process of mutation and selection. Mutations that result in the retention of the structural and biochemical functions of the viral proteins while simultaneously destroying antigenic determinants previously recognized by the adaptive immune system. Thus, a large pool of sequence variants is available for selection because the viral RNA-directed RNA polymerase lacks an editing function. This selection for minor variations in HA sequence has been termed antigenic drift. While this drift is sufficient to allow the virus to evade a robust adaptive immune response each flu season, it also may limit the ability of the virus to develop highly virulent variants during transmission within a particular host species.
In contrast, the emergence of pandemic strains has been associated with major HA sequence variations—antigenic shift—which appear to occur when a single host cell is co-infected with different viral strains resulting in virions that contain a variety of new assortments of the eight viral segments derived from different source viruses. It has been hypothesized that reassortment of genome segments may occur in species, like pig, with cells that present sialic acid with both the avian alpha 2,3 and human alpha 2,6 linkages. This could provide a mechanism for one viral clade to evolve through antigenic drift in one species where it develops the characteristics of a highly virulent strain for another species before crossing the species barrier following an antigenic shift event.
Influenza information management
Clearly, a detailed understanding of the interactions between virus and host would not only help us to understand the emergence of disease outbreaks, but also facilitate the development of improved diagnostics, therapeutics and vaccines to prevent and control influenza infection. A resource that goes beyond traditional bioinformatics is necessitated, and, if well constructed, would positively impact disparate fields in public health, molecular biology, life science information management and clinical studies. We aimed to create such a resource.
Many national and international health organizations have invested substantial resources in the support of research focused on improving our understanding of the pathogenesis of human infectious diseases. To bring together information from this valuable research, the National Institute of Allergy and Infectious Diseases recently funded the development of eight Bioinformatics Resource Centers for Biodefense and Emerging/Re-emerging Infectious Diseases (BRCs; http://www.brc-central.org/) focused on Category A–C pathogens (8). The BioHealthBase BRC is responsible for supporting data related to a select subset of these pathogens including influenza virus. The BioHealthBase BRC has assembled and integrated a variety of different types of data related to influenza virus, including gene and protein structure and function, sequence variation and immunological epitope information. In this manuscript, we describe the use of the BioHealthBase BRC to investigate the determinants of virulence in variant strains of avian H5N1 clade viruses, which are of special concern as a potential source for the next human pandemic strain.
DESCRIPTION
As of August 2007, information about ∼13 000 influenza virus strains is available at the BioHealthBase BRC. The BioHealthBase has been built upon a comprehensive foundation of gene and protein structure and function data from numerous external sources, including the NCBI, UniProt and the Immune Epitope Database (IEDB) (http://www.immuneepitope.org) (9) (Supplementary Figure 1A). The BHB support team derives and integrates novel data through the application of predictive bioinformatics algorithms and custom BHB-developed pipelines to primary sequence and annotation data for the pathogens under study. These data include immune epitopes, protein and RNA structures and protein localizations and genome sequence variations (Supplementary Figure 1B). The integration of available external data with information derived from computational prediction algorithms and manual curation provides a comprehensive framework to address scientific issues related to pathogen virulence.
In order to further understand the complexities of host–pathogen interactions, the BioHealthBase has contributed to the development of a comprehensive influenza life cycle within the Reactome database (10) project and is currently assisting in the completion of the influenza life cycle pathway details. A complete representation of the biological processes and molecular interactions necessary for viral replication and the host response to infection can be used for predicting targets for antiviral drugs and for determining the nature of virulence associated with protein sequence variants.
Scientific use cases: the Guangxi/35 example
To drive development of the BioHealthBase system, we have utilized scientific use cases to help define relevant data types, storage and query function and informatics processing workflows. For example, in 2005, Li et al. (11) described an analysis of H5N1 isolates obtained from healthy ducks in southern China, which varied in their ability to cause lethal infections in mice, with A/duck/Guangxi/22/2001 (DkXi22) being relatively avirulent and A/duck/Guangxi/35/2001 (DkXi35) being highly virulent. Using reverse genetic approaches, they found that virulence was partly dictated by the presence of Asn instead of Asp at position 701 of the PB2 protein. However, difference in other viral proteins, including NS1, also appeared to be involved. Utilizing the sequence data within the BioHealthBase, we will examine the additional causes of virulence of the DkXi35 strain.
Sequence search
We begin by utilizing the sequence search page specifically tailored for influenza virus-related data to examine these two strains in greater detail. Links to this search page are found along the upper left side of the BioHealthBase webpage. Simple keyword searches or advanced searches based on specific sequence annotation features (Figure 1A) may be performed on the influenza sequence search page. To find sequence records related to the DkXi35 strain, we searched for influenza A virus sequences of subtype H5N1 isolated from an avian host in China during the year 2001. The search page is also capable of excluding particular records by subtype, host, country and date range if necessary.
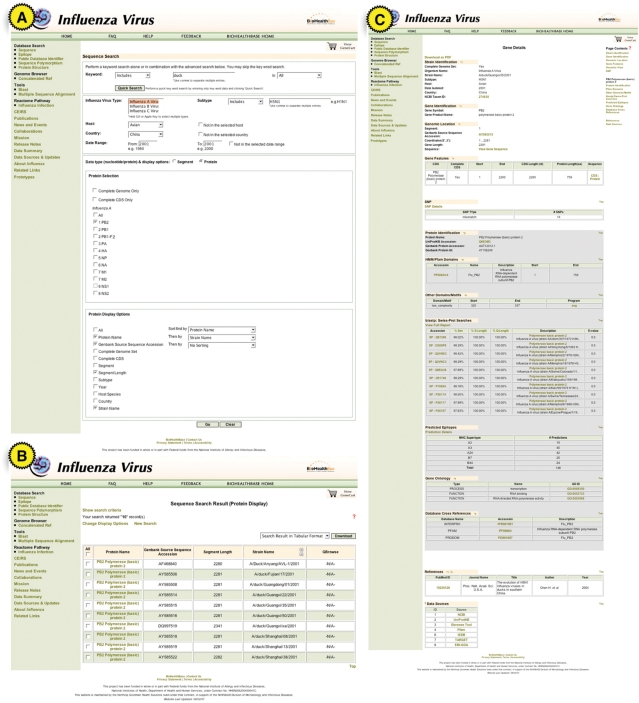
Figure 1.
Query access to annotation data. Screenshot of use case query/search page (A), summary results (B) and gene details for PB2 Quangxi/35 (C) are shown. Query constraint and report options and annotation details are described in more detail in the text.
We now turned our attention to the PB2 proteins of the selected strains. By selecting the protein data type, the sequence search page enables the selection of one or more proteins as well as limiting a search to full-length sequences or sequences belonging to a completely sequenced genome. By default searches include partial and full-length sequences and are not restricted to complete genome sets. Since we are only interested in full-length protein PB2 records, we select the full-length CDS option. We then select what sequence features to display in the search results and how the results records should be ordered (e.g. sort by strain name then segment).
The search returns 10 PB2 results including the DkXi22 and DkXi35 PB2 proteins (Figure 1B). By selecting one or more of the search results one is able to perform a variety of actions including downloading search results, or selected sequences in GFF3 or FASTA format or adding the sequences to a GeneCart (see later) for further analysis. Following the link from a record's gene symbol or protein name allows us to view the details of a particular sequence record. The Gene Details page contains all of the annotation integrated from external sources or computed internally for the selected sequence (Figure 1C). In the case of DkXi35, the annotation feature of particular interest is the single nucleotide polymorphism (SNP) annotation. For each gene (e.g. PB2) and species subtype (e.g. avian H5N1) a consensus sequence is computed. Each sequence is then compared to the consensus sequence and polymorphisms are identified using custom perl scripts. In summary, our analysis yielded 14 nt substitutions in the DkXi35 strain's PB2 gene, in comparison with the avian H5N1 consensus.
Sequence analysis using GeneCart
The BioHealthBase can save search results to a temporary workspace or ‘GeneCart’ for further analysis. In our use case, we save the DkXi22, DkXi35 and related PB2 sequences to the GeneCart. Additional sequence records derived from other searches can also be added. The GeneCart augments the sequence search capability of the BioHealthBase by allowing the assembly of disparate sets of sequences, which would be difficult to gather using a single search alone.
Once sequences have been added to the GeneCart they may be downloaded in FASTA or GFF3 format. The real power of the GeneCart is the ability to seamlessly perform BLAST analysis or multiple sequence alignment on one or more of the sequences in the GeneCart. For our analysis, we are interested in aligning the PB2 sequences as displayed in Figure 2A as well as the NS1 sequences as shown in Figure 2B. Multiple sequence alignment is performed using the MUSCLE (12) algorithm. Navigating to the multiple sequence alignment tool page from the GeneCart automatically populates the selected sequences into the sequence field for quicker alignments.
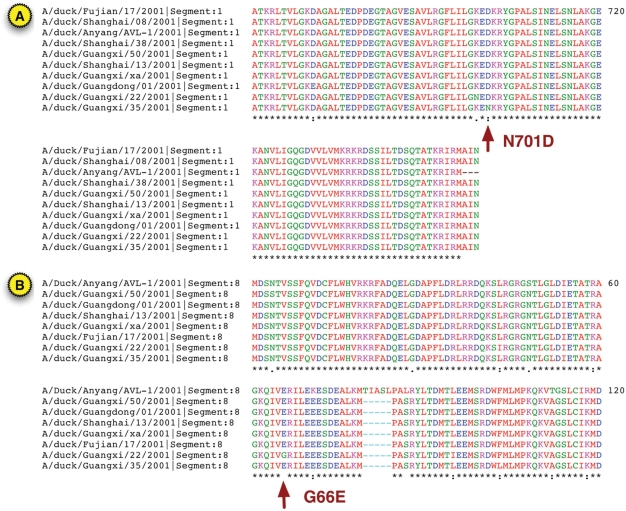
Figure 2.
Protein sequence alignment. The PB2 (A) and NS1 (B) protein sequences from all 10 influenza A virus H5N1 duck sequences isolated in China during 2001 were aligned separately using the BioHealthBase implementation of the MUSCLE algorithm. Only the PB2 protein region from aa661 to aa759 and the NS1 protein region from aa1 to aa120 are shown. The key N701D substitution found to affect virulence in DkXi35 PB2 and the G66E difference between DkXi22 and DkXi35 in the NS1 RNA-binding domain are highlighted.
Sequence feature visualization
In addition to the textual view of the Gene Details page sequences in the BioHealthBase can also be viewed in a 2D genome browser based on the GBrowse application (13). Sequence feature annotations are contained in ‘tracks’ that may be customized for viewing by turning them off or on or by re-configuring them. A user can also upload personal tracks of formatted annotation data. In our case, we can see that the NS1 segment contains 7-nt substitutions in comparison with the avian H5N1 consensus sequence, and two of these substitutions (colored in red) reflect amino acid changes that overlap with NetCTL (14) predicted T-cell epitopes of the human HLA A2 supertype (Figure 3).
Figure 3.
Genome structure view of DkXi35 segment 8. The RNA segment encoding the NS1 protein from A/duck/Guangxi/35/2001, the sequence variations versus the avian H5N1 consensus sequences and A2 supertype epitopes predicted by the NetCTL algorithm are shown.
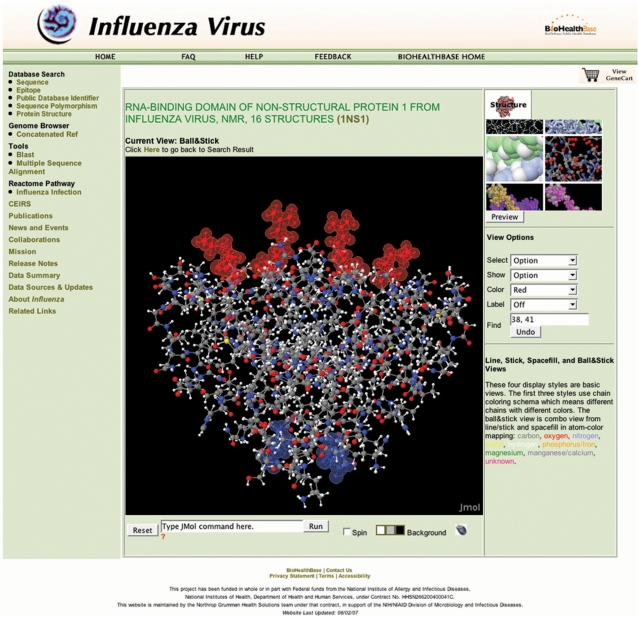
Protein structural analysis
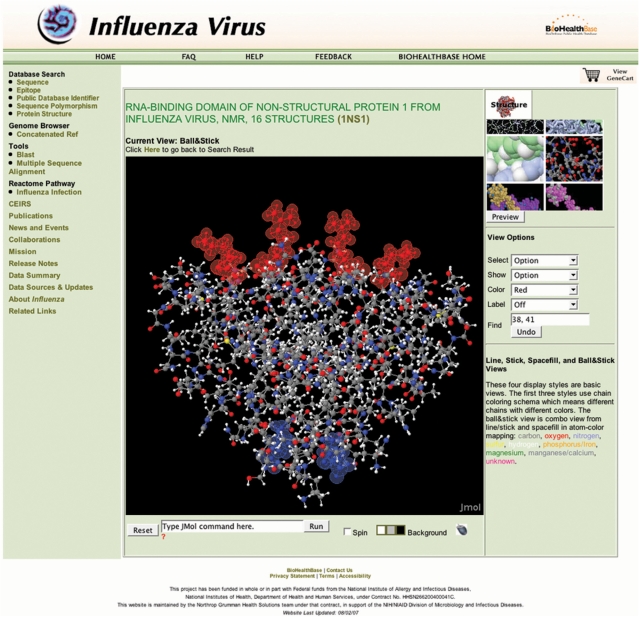
Finally, the BioHealthBase was used to visualize the physical relationship between the amino acid sequence variations in NS1 and the known functional regions of the protein using a 3D protein structure visualization window accessible through the left-hand menu. Proteins can be viewed in this tool in a variety of display formats (e.g. ball-and-stick, space-filling, ribbon) and different structural and functional regions highlighted. The viewer is based on a custom Jmol (http://www.jmol.org) implementation loaded with data from the Protein Database (15). In this use case, we mapped the amino acid variations from DkXi35 NS1 onto the structure determined for the RNA-binding domain of the NS1 protein from the A/Udorn/307/1972 isolate (Figure 4). From this analysis, it is clear that the amino acid sequence variation found in this region of the DkXi35 NS1 protein (G66E) is structurally distinct from the key RNA contact residues (aa38 and aa41), and is well outside of the RNA-binding pocket. Thus, this comparative analysis between sequence polymorphic variations and protein structural regions suggests that it is unlikely that the G66E variation influences virulence by affecting NS1–RNA interactions.
Figure 4.
3D Protein structure visualization. A ball-and-stick representation of the RNA-binding domain of the NS1 protein dimer from the A/Udorn/307/1972 (PDB ID 1NS1) is shown. RNA-binding residues aa38 and aa41 are highlighted in red, while the single amino acid difference between DkXi22 and DkXi35 at aa66 is highlighted in blue. RNA binds parallel to the plane on top of the red RNA-binding residues.
CONCLUSIONS
The BioHealthBase BRC provides a portal to a comprehensive range of biological data related to influenza virus physiology and pathogenesis. While several public database resources provide focused data sets about influenza virus isolates (e.g. sequence records), the BioHealthBase emphasizes the integration of data from public resources together with data derived from various analysis and prediction algorithms, allowing researchers to explore hypotheses using bioinformatics approaches before heading into the laboratory. The BioHealthBase places significant emphasis on supporting data related to host–pathogen interactions in order to gain a better understanding of the nature of virulence and host range, and the impact of sequence variation on these phenomena. Current plans for future enhancements of the BioHealthBase BRC resource include the ability to construct phylogenetic trees based on sequence relationships, the definition of functional sequence features in influenza proteins and their availability for display in both the genome browser and the 3D protein structure visualization module, and the support for surveillance and research data produced by the Centers of Excellence for Influenza Research and Surveillance program recently funded by NIAID (http://www3.niaid.nih.gov/research/resources/ceirs/).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the Reactome database project team, especially Marc Gillespie, Peter D’Eustachio and Lincoln Stein for their assistance with the development and deployment of the influenza life cycle representation. We also thank Aihui Wang, Bjoern Peters, Gillian Air, Feng Luo and Valentina Di Francesco for advice on various components of the BioHealthBase. The BioHealthBase Bioinformatics Resource Center has been wholly funded with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. N01-AI40041. Funding to pay the Open Access publication charges for this article was provided by NIH Contract No. N01-AI40041.
Conflict of interest statement. None declared.
REFERENCES
- 1.Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerg. Infect Dis. 2006;12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patterson KD, Pyle GF. The geography and mortality of the 1918 influenza pandemic. Bull. Hist. Med. 1991;65:4–21. [PubMed] [Google Scholar]
- 3.Palese P, Shaw ML. Orthomyxoviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. 5th. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 4.Ito T, Kawaoka Y. Host-range barrier of influenza A viruses. Vet. Microbiol. 2000;74:71–75. doi: 10.1016/s0378-1135(00)00167-x. [DOI] [PubMed] [Google Scholar]
- 5.Kuiken T, Holmes EC, McCauley J, Rimmelzwaan GF, Williams CS, Grenfell BT. Host species barriers to influenza virus infections. Science. 2006;312:394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- 6.Guillot L, Le Goffic R, Bloch S, Escriou N, Akira S, Chignard M, Si-Tahar M. Involvement of Toll-like receptor 3 in the immune response of lung epithelial cells to double-stranded RNA and influenza A virus. J. Biol. Chem. 2005;280:5571–5580. doi: 10.1074/jbc.M410592200. [DOI] [PubMed] [Google Scholar]
- 7.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus ADME, Fouchier RAM. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 8.Greene JM, Collins F, Lefkowitz EJ, Roos D, Scheuermann RH, Sobral B, Stevens R, White O, Di Francesco V. National Institute of Allergy and Infectious Diseases Bioinformatics Resource Centers: New Assets for Pathogen Informatics. Infect. Immun. 2007;75:3212–3219. doi: 10.1128/IAI.00105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters B, Sidney J, Bourne P, Bui H-H, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Chen H, Jiao P, Deng G, Tian G, Li Y, Hoffmann E, Webster RG, Matsuoka Y, et al. Molecular basis of replication of Duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 2005;79:12058–12064. doi: 10.1128/JVI.79.18.12058-12064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, et al. The generic genome browser: a building block for a Model Organism System Database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen MV, Lundegaard C, Lamberth K, Buus S, Brunak S, Lund O, Nielsen M. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur. J. Immunol. 2005;35:2295–2303. doi: 10.1002/eji.200425811. [DOI] [PubMed] [Google Scholar]
- 15.Westbrook J, Feng Z, Chen L, Yang H, Berman HM. The Protein Data Bank and structural genomics. Nucleic Acids Res. 2003;31:489–491. doi: 10.1093/nar/gkg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.