Abstract
EuroPhenome (http://www.europhenome.org) and EMPReSS (http://empress.har.mrc.ac.uk/) form an integrated resource to provide access to data and procedures for mouse phenotyping. EMPReSS describes 96 Standard Operating Procedures for mouse phenotyping. EuroPhenome contains data resulting from carrying out EMPReSS protocols on four inbred laboratory mouse strains. As well as web interfaces, both resources support web services to enable integration with other mouse phenotyping and functional genetics resources, and are committed to initiatives to improve integration of mouse phenotype databases. EuroPhenome will be the repository for a recently initiated effort to carry out large-scale phenotyping on a large number of knockout mouse lines (EUMODIC).
INTRODUCTION
With an extensive knowledge of the gene content of mammalian genomes becoming a reality through the completion of a number of genome sequences including mouse (1), focus has shifted to the study of gene function in these organisms. The aim in this postgenomic era is to link genomic and phenotype information systematically to allow a deeper understanding of the processes leading from genomic changes to altered phenotype and disease. Mouse mutants represent one of the most powerful tools in this endeavour (2). To facilitate these aims, a number of projects are underway ranging from the production of large collections of mouse point and knockout mutants (3,4) through to the establishment of large-scale phenotype characterization projects that aim to provide a phenotype assessment of these lines (5) (EUMODIC—http://www.eumodic.eu). The ultimate challenge will be to interpret this data using computational methods.
A number of critical steps must be achieved for the comprehensive analysis and interpretation of this data to be possible. First, phenotype data on both normal inbred strains and mutant strains must be collected in community databases with open access. Second, the phenotype data must be generated using comprehensive phenotyping platforms that provide standardized methods, so that the results can be compared between laboratories and across time (5). Third, structured descriptions of the phenotypes must be used to allow the data to be interpreted in a consistent manner (6–8).
EMPReSS (European Mouse Phenotyping Resource for Standardized Screens) (5,9) is the product of Eumorphia (http://www.eumorphia.org), the largest programme to date to develop standardized phenotyping protocols. EMPReSS is a comprehensive database of validated Standard Operating Procedures (SOPs) for screens to determine the phenotype of a mouse. It incorporates 96 SOPs that cover all of the main body systems including: clinical chemistry, hormonal and metabolic systems, cardiovascular, allergy and infection, sensory function, neurological and behavioural function, cancer, and bone and cartilage systems. In addition, there are generic SOPs for histology, necropsy, pathology and gene expression. EMPReSS is a platform of individual tests, but these can also be grouped together into phenotyping pipelines.
EuroPhenome (http://www.europhenome.org) was instigated as an online mouse phenotyping resource to store baseline data generated from the application of EMPReSS SOPs. Data was collected by individual work packages in the Eumorphia project for purposes of SOP validation and to provide baseline information against which phenotypes of experimental animals could be compared. It currently includes data from 24 EMPReSS SOPs, representing the measurement of 132 parameters across four inbred mouse strains in up to four different laboratories.
EUROPHENOME
EuroPhenome is a mySQL relational database (http://www.mysql.com/) providing baseline data on four inbred mouse strains (C57BL/6J, C3H/HeBFeJ, BALB/cByJ and 129/SvPas). Data were obtained by researchers working in the Eumorphia project. The data are subdivided into five phenotyping domains corresponding to body systems (see Table 1).
Table 1.
Summary of datasets included in Europhenome—August 2007
| Phenotyping domain | SOP name | Number of parameters measured |
|---|---|---|
| Behaviour and cognition | Open field | 7 |
| Grip strength | 3 | |
| Y-maze | 4 | |
| Acoustic startle and pre-pulse inhibition specific to sensory systems | 3 | |
| Tail flick | 1 | |
| Clinical and haematology | Differential blood count | 2 |
| Clinical chemistry parameters | 27 | |
| Haematology tests | 7 | |
| Hormonal and metabolic systems | Metabolic cages | 1 |
| Dexa scan analysis | 5 | |
| IPGTT (intra-peritoneal glucose tolerance test) | 2 | |
| Cold test | 2 | |
| Meal tolerance test | 4 | |
| Oral glucose tolerance test | 2 | |
| Cardiovascular | Non-invasive blood pressure and heart rate | 2 |
| Invasive blood pressure | 3 | |
| Invasive left-ventricular haemodynamics | 5 | |
| Surface electrocardiography | 5 | |
| Echocardiography | 20 | |
| Allergy and infectious diseases | FACS analysis of peripheral blood cells | 9 |
| Determination of immunoglobulin concentration in serum | 6 | |
| Quantification of TNFa production by PAMP stimulated macrophages | 7 | |
| Infection of proteose peptone macrophages with Listeria monocytogenes | 1 | |
| Quantitative measurement of iNOS activity after macrophage stimulation | 4 |
Navigating EuroPhenome
The EuroPhenome data browser utilizes PHP and AJAX and gives the user a variety of ways to visualize and search the data, allowing assessment of inter-strain, -gender and -laboratory variation for the various SOPs. The web-accessible data browser (http://www.europhenome.org/browser) allows users to browse through the data via the SOP. SOPs are presented in a tree structure on the left-hand side of the browser. SOPs are clustered under five phenotyping domains, corresponding to the classification in EMPReSS. Opening up a folder corresponding to one of these domains reveals a list of SOP names. Clicking on an SOP name opens a summary page in the main frame of the browser that lists the parameters measured under the selected SOP and mean, median and variation measures over the entire dataset for each parameter (Figure 1). Users can also link directly to the description of the SOP in EMPReSS, to similar measurements in the Mouse Phenome Database (10), and download the data to Excel format, allowing them to carry out their own analyses of the data and to combine it with their own in-house data. Clicking on an individual parameter reveals a more detailed breakdown of parameter means and SDs broken down by age and sex. These values are also represented graphically for easy visual comparison (Figure 2). The left-hand panel also provides a link to Phenostat, which can be used for statistical analysis of datasets (11).
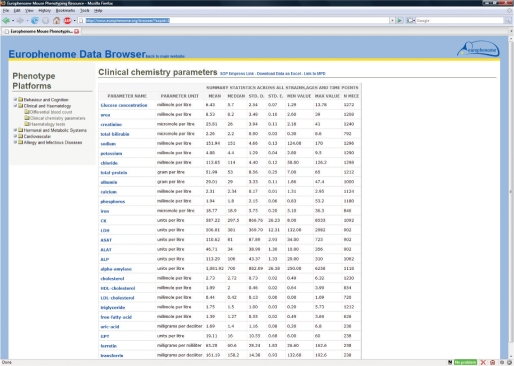
Figure 1.
View of the summary data display for EuroPhenome. The left-hand panel contains the tree-like browser that allows users to search for data derived from individual SOPs in a structured manner. The main panel contains a table displaying Parameter Name, Parameter Measurement Unit, Mean, Median, SD, Standard Error, Minimum and Maximum values, and number of mice measured. Links are also provided to the EMPReSS SOP, download the data in Excel format, and link to the corresponding MPD dataset. Clicking on the name of an individual SOP in the left-hand panel takes the user to a more detailed view (Figure 2). The example shown is for the SOP ‘Clinical chemistry parameters’.
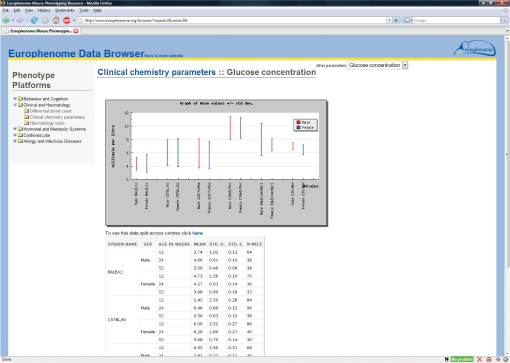
Figure 2.
View of the detailed data display for an individual parameter in EuroPhenome. The graphical display at the top of the main panel illustrates mean values of the parameter for each mouse strain tested, separated into male and female. Error bars represent SDs. The table below gives Mean, SD, Standard Error and mouse numbers for each strain subdivided into sex and, where appropriate, age groups. Clicking on the link ‘To see this data split across centres click here’ expands this table to show results obtained in the different Eumorphia laboratories that carried out the test. The pull-down menu at the top right allows navigation between the different parameters measured by a given SOP, where appropriate.
EuroPhenomeMart
In addition to this browsing interface, we have also implemented a MartView/BioMart (http://www.biomart.org/) interface to the database, which can be accessed from a link on the left-hand panel of the browser. This allows export of selected fields to HTML pages, and in comma or tab separated formats.
Implementation of web services in EuroPhenome
Open standard web services technology such as Simple Object Access Protocol (SOAP, http://www.w3.org/TR/soap) and Web Service Definition Language (WSDL, http://www.w3.org/TR/wsdl) enable programmers to build complex applications without the need to install and maintain the database, and facilitates integration and interoperability between bioinformatics applications and the data they require.
Web services have been implemented in EuroPhenome to expose its data in a programmatically accessible manner. We are currently using RPC/Encoding style WSDL and will be providing Document/Literal style soon, as recommended by the Web Services Interoperability Organization (WS-I, http://www.ws-i.org). The URL for accessing the EuroPhenome WSDL file is http://www.europhenome.org/europhenome.wsdl.
EMPRESS—RECENT DEVELOPMENTS
The original implementation of the EMPReSS SOP site is described in Green et al. (9) and its scientific genesis in (5). The implementation of EMPReSS has subsequently been redeveloped and the browser is now a web 2.0 resource combining JAVA, XML, AJAX and PHP for the visualization and searching of the SOPs. This functionality now enables bioinformaticians to access the data in the eXist XML database (http://exist.sourceforge.net/) via web services. The EMPReSS browser has been extended to allow users to select data generated from SOPs in the two EMPReSSslim phenotyping pipelines (see below). The pipelines are comprehensive sequences of SOPs that are being utilized for high-throughput phenotyping in the EUMODIC project. The interface also allows direct access to available data generated using a particular SOP in EuroPhenome (Figure 3).
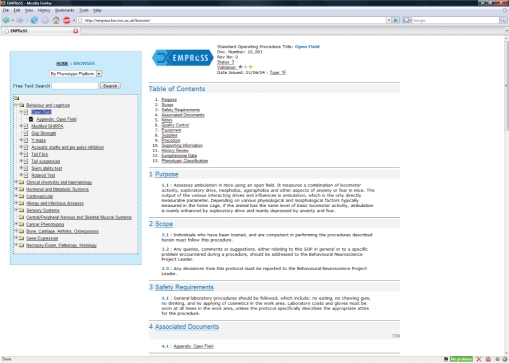
Figure 3.
View of the SOP display in the EMPReSS browser. The tree-like display in the left-hand panel provides for easy navigation to individual SOPs, which are classified under the body system they investigate. The right hand panel displays the SOP itself. The SOP document is subdivided into 11 sections as described previously (9) plus an additional section providing a link to EuroPhenome. The view also provides status information about the SOP including a unique document ID, revision number (previous versions of SOPs are retained as appendices to the current version; a new version is only recorded is there has been a substantive change), review status and the validation level of the SOP [see (9) for details].
The EMPReSS SOPs have also been annotated with high level Mammalian Phenotype (MP) ontology terms (8) and an ontology tree browser interface developed to facilitate the identification of the annotated SOPs. This annotation currently uses top-level MP terms, but we aim to develop it further in depth and diversity.
The ontology tree browser utilizes the Ontology Lookup Service (OLS) (http://www.ebi.ac.uk/ontology-lookup/) (12) at the EBI to provide the ontology data to dynamically generate an AJAX-driven JavaScript and DHTML ontology tree menu. This allows the user to search through the SOPs by specific ontology term. This system works in combination with Memcached (http://www.danga.com/memcached/), a high-performance, distributed memory object caching system, to negate any potential slow networking issues inherent in web service access. Implementing the OLS system allows the flexibility of selecting any of the 48 currently stored ontologies to dynamically generate an ontology tree and thus search through the SOPs using those terms. It also facilitates an AJAX-driven autocompletion search functionality, providing an easy way to hunt for a specific term within an ontology and use it to search within the SOPs. The results of a search are provided as a list, enabling the user to load the SOP of interest, and it also provides a link out to the MGI database of mutant lines annotated with the corresponding MP term.
FUTURE DIRECTIONS
The major future objective for EuroPhenome is to move from the collection of phenotyping data on inbred strains to mutant strains. EuroPhenome will be the primary repository and query tool for data generated from the EUMODIC project, which will phenotype up to 650 knockout lines generated by the EUCOMM project (http://www.eucomm.org). EUMODIC has developed a phenotyping screen called EMPReSSslim, which is structured for comprehensive, primary, high-throughput phenotyping of large numbers of mice. The primary phenotype assessment using EMPReSSslim will be undertaken in four large-scale phenotyping centres at the GSF, Germany; ICS, France; MRC Harwell, UK and the Sanger Institute, UK. Data generated in the centres will be submitted to EuroPhenome as XML files. It is envisaged that the EuroPhenomeXML Schema could be used more generally for the exchange of phenotyping data.
EuroPhenome is a core member of the Mouse Phenotype Database Integration Consortium (MPDIC) (13) and CASIMIR (Coordination and Sustainability of International Mouse Informatics Resources; http://www.casimir.org.uk/). As such, one of its primary aims over the next 1–2 years is to integrate with many other data sources including Ensembl (http://www.ensembl.org), MGI (http://www.informatics.jax.org/), RIKEN (http://www.gsc.riken.go.jp/Mouse/), EUCOMM (http://www.eucomm.org), EURExpress (http://www.eurexpress.org) and ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) to enable intelligent querying of the data. We already support some integration with the Mouse Phenome Database (see above) and have shared our SOPML with RIKEN. To develop this integration, we will in part utilize existing technologies such as BioMart and DAS, but will also investigate alternative methods. Of particular importance in this effort will be the development of new standards, as described elsewhere (13). Particular areas for development include:
Standards for phenotype data transfer. Building on the XML schema being developed for data capture within EUMODIC, and on other initiative to describe phenotypes in XML, such as PhenoXML (http://www.fruitfly.org/~cjm/obd/pheno-xml-xsd.html) a standard format will be developed to facilitate the exchange of phenotype data between sites, enabling integrated analysis and searching of phenotype data across different sites.
Standards for describing phenotyping procedures. In collaboration with members of the MPDIC we are currently developing an enhanced version of SOPML, which is currently used to store phenotyping SOPs in EMPReSS, to make it more descriptive and include ontological mark-up. This is intended to act as an international standard for the description, storage and interchange of Phenotyping Procedures.
Using ontologies to describe phenotyping data. Data from phenotyping experiments require to be described in a value-neutral way, i.e. they should not be annotated as ‘abnormal’ until proper controls and data filtering methods have been applied. They also need to be described at a more detailed level than is available using MP (8). While taking note of future developments in MP, we will also develop the use of combinatorial approaches to phenotype description that include descriptions of the underlying experimental protocol (6,7) combined with recent developments in the PATO ontology (http://www.bioontology.org/wiki/index.php/PATO:Main_Page) and integrate these into the description frameworks mentioned above to allow for transfer of semantic information about experimental data held in EuroPhenome.
A longer-term aim is to open up EuroPhenome-EMPReSS to the broad community as a repository for phenotype data via a linked submission procedure that would acquire information about phenotyping procedure, data, and ontological markup.
ACKNOWLEDGEMENTS
We thank Simon Greenaway for initial work on the EuroPhenome database schema and Aadya Shukla for coordinating the collection of raw data and for a preliminary draft of the database. We also thank the members of the EUMORPHIA consortium for their work in drafting the SOPs and generating the baseline data. Finally, we thank Hilary Gates and Mandy Studley for outstanding support during the development of these projects. European Commission (QLG2-CT-2002-00930, LSHG-CT-2006-037188); UK Medical Research Council. Funding to pay the Open Access publication charges for this article was provided by UK Medical Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 2.Brown SDM, Hancock JM, Gates H. Understanding mammalian genetic systems: the challenge of phenotyping in the mouse. PLoS Genet. 2006;2:e118. doi: 10.1371/journal.pgen.0020118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordes SP. N-ethyl-N-nitrosourea mutagenesis: boarding the mouse mutant express. Microbiol. Mol. Biol. Rev. 2005;69:426–439. doi: 10.1128/MMBR.69.3.426-439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins FS, Rossant J, Wurst W. International Mouse Knockout Consortium. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 5.Brown SDM, Chambon P, Hrabé de Angelis M, Eumorphia Consortium EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nat. Genet. 2005;37:1155. doi: 10.1038/ng1105-1155. [DOI] [PubMed] [Google Scholar]
- 6.Gkoutos GV, Green ECJ, Mallon A.-M, Hancock JM, Davidson D. Using ontologies to describe mouse phenotypes. Genome Biol. 2004;6:R8. doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gkoutos GV, Green ECJ, Mallon A.-M, Hancock JM, Davidson D. Building mouse phenotype ontologies. Pac. Symp. Biocomput. 2004;9:178–189. doi: 10.1142/9789812704856_0018. [DOI] [PubMed] [Google Scholar]
- 8.Smith CL, Goldsmith CA, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2004;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green ECJ, Gkoutos GV, Lad HV, Blake A, Weekes J, Hancock JM. EMPReSS: European mouse phenotyping resource for standardised screens. Bioinformatics. 2005;21:2930–2931. doi: 10.1093/bioinformatics/bti441. [DOI] [PubMed] [Google Scholar]
- 10.Bogue MA, Grubb SC, Maddatu TP, Bult CJ. Mouse Phenome Database (MPD) Nucleic Acids Res. 2007;35:D643–D649. doi: 10.1093/nar/gkl1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reuveni E, Carola V, Banchaabouchi MA, Rosenthal N, Hancock JM, Gross C. Phenostat: visualization and statistical tool for analysis of phenotyping data. Mamm. Genome. 2007;18 doi: 10.1007/s00335-007-9042-4. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Côté RG, Jones P, Apweiler R, Hermjakob H. The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinformatics. 2006;7:97. doi: 10.1186/1471-2105-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mouse Phenotype Database Integration Consortium. Integration of mouse phenome data resources. Mamm. Genome. 2007;18:157–163. doi: 10.1007/s00335-007-9004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]