Abstract
Arabidopsis thaliana Protein Interactome Database (AtPID) is an object database that integrates data from several bioinformatics prediction methods and manually collected information from the literature. It contains data relevant to protein–protein interaction, protein subcellular location, ortholog maps, domain attributes and gene regulation. The predicted protein interaction data were obtained from ortholog interactome, microarray profiles, GO annotation, and conserved domain and genome contexts. This database holds 28 062 protein–protein interaction pairs with 23 396 pairs generated from prediction methods. Among the rest 4666 pairs, 3866 pairs of them involving 1875 proteins were manually curated from the literature and 800 pairs were from enzyme complexes in KEGG. In addition, subcellular location information of 5562 proteins is available. AtPID was built via an intuitive query interface that provides easy access to the important features of proteins. Through the incorporation of both experimental and computational methods, AtPID is a rich source of information for system-level understanding of gene function and biological processes in A. thaliana. Public access to the AtPID database is available at http://atpid.biosino.org/.
INTRODUCTION
At the cellular level, a network of molecular interactions is representative of life. Cellular transport such as the movement of molecules and macromolecules from one location to another within cells and the formation of complex molecular structures make the properties of the network more intricate. However, all of this apparent complexity can be systematically illustrated as a simple interaction network, particularly through an understanding of protein–protein interaction (PPI) networks.
The collection of all protein interactions in an organism is typically referred to as an interactome (1). PPIs are fundamental to virtually every aspect of cellular function (2). PPI provides useful information of functional linkage between interacting partnerships within cells (3). Therefore, PPI can help to reveal signal transductions (4,5), post-translational modifications and developmental processes (6). In addition, it can serve to aid in the identification of novel regulatory components and pathways, and provide a valuable approach to understand functional specificities at the molecular level.
Sequence-based annotation efforts have led to the identification of a number of cellular components, which can be regarded as a one-dimensional annotation. Accumulated information regarding interactions, and advancement of various high-throughput technologies make it possible to generate systematical, or two-dimensional annotations (7), such as interaction maps.
The past several years have witnessed an exponential increase in the amount of biological data, mainly due to the development and application of high-throughput technologies, including gene expression microarrays and mass spectrometry to characterize DNA, RNA and proteins. Currently, interactomes have been created for several model organisms, such as Saccharomyces cerevisiae (8,9), Drosophila melanogaster (10), Caenorhabditis elegans (11) and Homo sapiens, among others (12). In the plant kingdom, Arabidopsis thaliana has been widely employed as a model organism to elucidate important biological principles. In fact, several years ago the entire sequence of the Arabidopsis genome was reported, and most of its genes annotated (13). However, there are still 30% of these gene products yet to be characterized because sequence homology was not effective at assigning gene function. Only a few interactions of specific protein families in A. thaliana have been reported (14,15), however, an enhanced understanding of PPIs can suggest important future directions for researchers to study gene/protein relationships and functions.
Meanwhile, many experimental procedures have been developed to analyze PPIs, including biochemical methods [e.g. protein affinity chromatography (16–18), affinity blotting, coimmunoprecipitation and cross-linking (19–22)]; molecular biological methods [e.g. protein probing, the two-hybrid system (23–25) and phage display (26)] and genetic methods [e.g. the isolation of extragenic suppressors and synthetic mutants (27)]. High-throughput experimental techniques have enabled us to study PPIs at the proteome scale. This is achieved via systematic identification of physical interactions among all proteins in an organism. The ever-increasing volume of PPI data is becoming the foundation for new biological discoveries. However, these data are distributed in numerous sources and it has been confirmed that some data are noisy, data quality varies significantly, and data often cannot be verified against each other. Bioinformatics and computational approaches have been used to assess the reliability of high-throughput results and to gain confidence in published data (28). The methodology can also integrate raw data into useful information and provide experimentally testable hypotheses, thereby expanding our knowledge about new mechanisms in biological processes (29–32). Other computational prediction methods based on known protein structural interactions can also be useful to analyze large-scale PPI rules. This prediction methodology evaluates interaction rules among complete genomes using protein structural interactome maps (33). Consequently, numerous researches using computational methods have been carried out to investigate gene and protein functions, PPIs and gene regulation relationships(34–39). These approaches have been applied to interactomes of H. (40), S. cerevisiae (41), C. elegans (37), Plasmodium falciparum (42,43), among other organisms.
However, rapidly increasing amounts of biological data generated from genome-wide and proteome technologies on modern biochemistry and molecular biology need to be well stored, compararable, organized and accessible. An appropriate repository and maintenance system for these data can facilitate future data mining and functional investigations.
AtPID was developed using A. thaliana as the model system for a comprehensive resource of PPIs. All data in AtPID were deposited from either manual text mining or bioinformatics predictions. This database contains 28 062 interaction pairs, of which 3866 involve 1875 proteins obtained from the literature and 800 pairs were from enzyme complexes in KEGG. In addition, bioinformatics predictions or literature surveys provided 5562 proteins with subcellular location information. Intuitive and user-friendly query interfaces have made all the features of AtPID easily accessible. This database provides invaluable resources for researchers to study PPIs and protein functions in Arabidopsis, data can also be used to address questions regarding gene functions and biological processes in other taxa. AtPID is a non-commercial public access database (http://atpid.biosino.org/) that provides data download services for standalone analyses or data mining, including protein interaction properties and other areas of interest in plant biology.
DATA SOURCES AND PROCESSING
Data resources for reconstruction of interaction network
The power and expressivity of any network lies largely in the data model used to represent molecular interactions. From a computational perspective, we applied uniform systematic benchmarks and statistical approaches to specifically train our PPI network for Arabidopsis. In addition, to assure data quality, we treated each resource separately as weighted features and reconstructed the PPI network through the proper integration of various protein interaction datasets according to the Naïve Bayesian Network theory. In this way, meaningful biological data is made available through AtPID. Here protein interaction data are generated in the following ways: experimental results are obtained from related papers in PubMed and other available databases; and data are made accessible from bioinformatics predictions. The details of interaction data generation are described below.
Manually collected protein interactions were extracted from not only thousands of published articles, but also IntAct (44), BIND (45) and TAIR databases (13). We deposited protein interaction data possessing physical evidence or experimental references related to the association between two proteins into AtPID. To ensure the reliability of these data, we also conducted a validation process. First, we mapped PPI collected from the literature lacking AGI locus identifiers to IPI (46) and removed symbols without a match. We applied uniform AGI symbols to proteins in AtPID for further analysis. We found 3866 PPI pairs involving 1875 proteins using this filtration process. Additionally, protein pairs in enzyme complexes were also inferred as a part of GSP based on the assumption that subunits in an enzyme complex have high functional association and potential physical interactions. Enzyme complexes from KEGG (47) were obtained to extract the intersection of interactions from text mining and complexes of enzymes directly garnered from the KEGG database. We subsequently used the decision tree to determine how many proteins belonging to an enzyme complex resulted in a less false positive and higher accuracy. Because many subunits or components of an enzyme complex are mapped from sequence similarity with other species or orthologs, we compared true protein interaction data to reduce noise and redundant information. Eventually, 800 unique pairs were obtained through enzyme complex after excluding the redundancies from the 3866 pairs via text mining. Consequently, a total of 4666 interaction pairs involving 2285 proteins were generated (Table 1). Such protein interaction resources, called GSP (Golden Standard Positive) are stored in AtPID and used to score the interaction network that assigns each predictive interaction pair with quantized measures.
Table 1.
Overview of GSP resources
| PPI Resources | Number of.PPI pairs | Number of proteins in PPI pairs | |
|---|---|---|---|
| GSP PPI | [1] Literatures from PubMed | 1259 | 740 |
| [2] InAct | 1528 | 677 | |
| [3] BIND | 1475 | 538 | |
| [4] TAIR | 1073 | 698 | |
| [1]∼[4] | 3866 | 1875 | |
| Protein complexes | [5] KEGG (enzyme complex) | 1700 | 856 |
| Total | [1]∼[5] | 4666 | 2285 |
[1] Manually collected protein interactions are extracted directly from thousands of published articles in PubMed. [2] InAct provides a freely available, open source database system for protein interaction data in EMBL-EBI. All interactions are derived from literature curation or direct user submissions. [3] BIND is a new resource to perform cross-database searches of available sequence, interaction, complex and pathway information. It integrates a range of component databases including Genbank and BIND, the Biomolecular Interaction Network Database. [4] TAIR provides ‘Tair Protein Interaction’ file by Matt Geisler at its FTP (ftp://ftp.arabidopsis.org/home/tair/Proteins/). [5] KEGG, a reference knowledge base linking genomes to biological systems and environments, provides resourceful enzyme complex information. [1]∼[4] After mapping various symbols to AGI, we found 3866 PPI pairs involving 1875 proteins with literature supports. [1]∼[5] combined with enzyme complexes from KEGG, the total number of GSP is up to 4666 involving with 2285 proteins.
For predicting PPIs in AtPID, we applied computational approaches, including conserved protein interactions (i.e. interologs) (48), gene expression data (49,50), genomic context (i.e. gene neighbor algorithm) (51,52), gene fusion (Rosetta Stone method) (53,54), phylogenetic profiles (55,56) and GO annotation. The optimized phylogenetic profiles were constructed and assessed using the method of Sun et al. (57). Orthologous PPIs in A. thaliana were obtained according to ortholog function transfer. Ortholog map files in Inparanoid (58) and DIP interaction data for other organisms were also collected (59). To infer Arabidopsis protein interactions, we mapped several model organismal (e.g. S. cerevisiae, D. melanogaster, C. elegans and H. sapiens) protein interaction data and orthologs to Arabidopsis. In addition, we used the atlas of Arabidopsis development microarray data (Acc.no: ME00319) from the TAIR database (13) to identify co-expressed genes.
Non-redundant proteins with GO annotation from the Gene Ontology Consortium were identified. These data were used to calculate the Shared Smallest Biological Processes (SSBP) value of each pair for all proteins employing GO annotation methods (40). Interacting proteins often function in the same biological process. Therefore, proteins involved in the same process are more likely to interact than proteins in distinct processes. Furthermore, proteins exhibiting high functional specificity are more likely to interact than proteins functioning in more comprehensive processes. Based on this assumption, we first identified all biological process terms shared by two proteins. Subsequently, we counted how many other proteins are assigned to each of the common terms and produced the shared biological process term with the smallest count (SSBP). In general, the smaller the SSBP count, the more specific the biological process term, and the greater the functional similarity between two proteins. In this way, we used SSBP to predict PPIs.
We also investigated the assumption that some of the operons contained within a particular organism may be conserved across other organisms based on the Gene Neighbors method. The conservation of an operon's structure provides additional evidence that genes within an operon are functionally coupled and are perhaps components of a protein complex or pathway.
Finally, we adopted the gene fusion method. The underlying assumption of the method is that if a composite protein is uniquely similar to two component proteins in another species, the component proteins are most likely to interact (53). Gene-fusion events were identified in complete genomes, based solely on sequence comparisons. These data enable the inference of functional associations among proteins.
The Bayesian Networks approach
The predictive datasets from such individual methods were integrated employing Naïve Bayesian Networks (40). The Bayesian Networks approach was used to integrate more than seven predictive data sources and to subsequently build a model to infer novel PPIs for Arabidopsis. The essence of the approach is to provide a mathematical rule, given some predictive evidence, to explain how to adjust the odds that a pair of proteins interacts, either in a true interaction instance (GSP) or correspondingly, in negative protein interactions, known as GSN (Golden Standard Negative). No direct information regarding the absence of specific protein interactions is available. However, protein localization data provides indirect evidence, given we assume that proteins in different cellular compartments do not interact (60). Hence, GSN values were constructed based on this assumption using subcellular localization data from the SUBA database (61). Individual likelihood ratios were easily calculated by counting the number of protein pairs with values that overlap with the GSP and GSN sets in the predictive dataset.
The confidence scores (LR) for each inferred PPI pair were the sum of the logarithmic form of all seven individual likelihood ratios from corresponding methods. The AtPID querying results page depicts the LR score from each method with open, partially or completely filled circles that indicate positive correlations with the confidence level of the interaction relationship. The detailed number of each predictive dataset is shown in Table 2 and all predictive datasets can be downloaded from the AtPID website.
Table 2.
Overview of the number of individual predictive dataset
| Number of predictive PPI pairs | Number of proteins in the PPI pairs | |
|---|---|---|
| O: Ortholog interaction datasets | 3045 | 1359 |
| G: Shared biological function:GO Ontology | 553 | 523 |
| E: Co-expression | 14 837 | 8024 |
| F: Gene fusion method | 6570 | 5671 |
| N: Gene neighbors method | 2008 | 1637 |
| P: Phylogenetic profile method | 15 723 | 8751 |
| D: Enriched domain pair | 2182 | 1288 |
| AtPIDa | 28 062 (putative PPI with GSP) | 12 506 |
| 23 396 (putative PPI without GSP) | 11 706 |
aThrough integration by Naïve Bays Network, AtPID achieved 28 062 PPI pairs with 23 396 pairs from prediction methods. There are seven individual datasets from various approaches, identified by O, G, E, F, N, P and D. The details of each method can be browsed on AtPID FAQ.
DATABASE CONTENTS AND USAGE
All of the information in AtPID is derived from expert curation and deliberate computation. The process of creating a release AtPID database begins with extracting the published and other relevant information from various databases (Figure 1). Automated and manual quality assurance procedures are administered to verify data completeness and consistency. If necessary, material in the development database is revised and a new database version is generated.
Figure 1.
The home page of AtPID.
Ortholog maps, domain attributes and network displays are developed with crosslinks to other relevant external resources (62). Following the final testing of data and the web server, the new database was made available via the public website. The latest release (14 July 2007) contains 28 062 PPI pairs involving 12 506 proteins. Of the PPI pairs, 23 396 pairs were inferred by the integration of several methods, while the other 3866 pairs involving 1875 proteins were manually curated from the literature and other 800 pairs were determined from enzyme complexes from KEGG. In addition to protein interaction data, we added subcellular location annotations to nearly 5000 proteins from SwissProt and SUBA databases (61,63) and popular prediction tools, including TargetP (64), Predotar (65) and MitoProtII (66), which can promote subproteome and protein function research.
AtPID spans roughly 41% of the estimated 30 480 peptides with interaction annotations in the Arabidopsis genome and reflects the labor-intensive nature of manual curation. Our future plans are to manually mine thousands of protein interactions to acquire information through bulk importation of data from other sources or experimental results. Thus increasing PPI information and power as a resource. PPI will also provide enhanced resource training data for reconstructing interaction networks with higher accuracy and larger coverage of the Arabidopsis genome. In turn, the database can aid users in querying more detailed information about interaction pathways or maps comprising of interesting protein attributes.
Practical applications of AtPID: querying interactions
The AtPID website can be browsed similar to an online library. The website's home page, depicted in Figure 1, features the ‘QUICK START’ main interaction querying box with links to each of the seven method theories used in AtPID, the AtPID database statistics, and announcements regarding the function of the website.
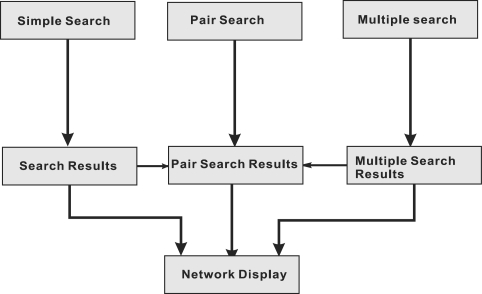
PPI query is the main function of AtPID, which makes available manually collected PPI data and predicted PPI through integrated data resources. Query flow is illustrated in Figure 2 and demonstrates how querying a protein name or protein pair on the query page accesses PPI information (http://atpid.biosino.org/query.php). AtPID allows several types of query keywords used by other databases, including UniProtKB/Swiss-Prot ID, TAIR AGI, Entrez Gene name, REFSEQ PROVISIONAL ID (NCBI) or International Protein Index (IPI) symbols. We defined three types of submissions. (i) ‘Simple search’ allows the user to submit a single protein. This search is appropriate when the user would like to know which other protein(s) have the highest probability of interacting with the protein of interest. The search results include the GSP and PPI predictions. (ii) ‘Pair search’ allows the user to submit a protein pair to ascertain if an interaction between two proteins has been documented. (iii) ‘Multiple search’ allows a user to query more than two proteins. A comma separate format is required to access an interaction network among multiple proteins. All returned pages inform the user of related protein annotations by text and graphs. For example, the user is interested in the HAP3A protein, which encodes a subunit of the CCAAT-binding complex and binds to the CCAAT box motif present in some plant promoter sequences. The ‘Search Results’ show a summary of the protein attributes in the first table, including the ‘Locus’, gene/protein symbol, the number of interactions (six from GSP and seven by inference), function description and database cross-references to Entrez, TAIR, IPI, UniProtKB/TrEMBL, UniParc and KEGG (Figure 3).
Figure 2.
An overview of querying flow.
Figure 3.
The AtPID interface of PPI Simple Search Results for the queried protein, HAP3A. There are generally three tables: (1) protein attributes with gene model descriptions and PPI summary, showing how many protein pairs can be predicted and the inferred interactions overlapped with GSP. (2) The second table represents interactants of the queried protein, HAP3A. Users can view what the experimental evidence is to support this consequence and link to corresponding literature(s). (3) The third table represents potential interactants of the queried protein, HAP3A, by Naïve Bays integration across the seven approaches. Each approach is graphically represented by circles. When the cursor is over a circle, it will display the corresponding score (LR) for the method.
The second table of the ‘Search Results’ presents inferred PPI pairs belonging to GSP listed with supporting evidence, including literature references from PubMed and experimental detection methods from text mining. Each interactant can be linked to a new ‘Pair Search Results’ window. The third table displays any potential interactant of HAP3A (Figure 3). Each of the seven prediction approaches is depicted by a letter acronym within a circle. When the user places the cursor over each circle, it displays the full name of the method. The circle under each method indicates the confidence strength for the predicted method and the related protein. The more the circle is filled, the more likely the pair of proteins is to interact. The corresponding score for the specific method is displayed when the cursor is held over the circle.
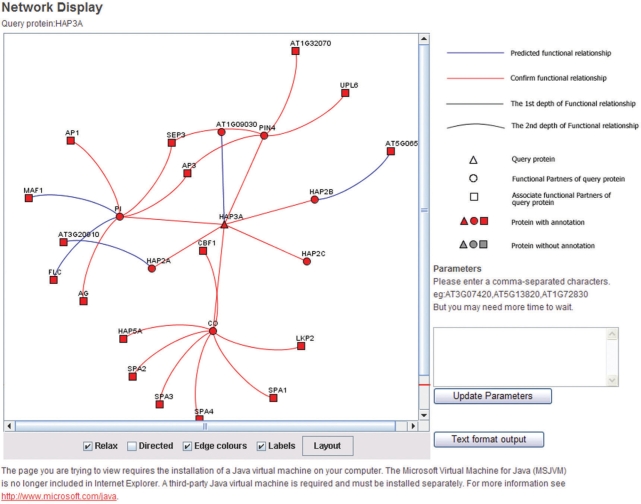
‘Network Display’ above the information table provides the link to a new window that displays the interaction network about HAP3A (Figure 4). In the ‘Network Display’ page, the query protein is represented as a triangle; the functional Partners of the queried protein are represented by a circle, derived from the first level displayed in the PPI network that directly linked to the query protein. The associated functional partners of the queried protein are shown as squares, derived from the second level in the network. A red node represents a protein with known function (i.e. annotated), whereas a gray node represents an unknown functional protein (i.e. without annotation). The line between each protein indicates the functional relationship; a red line infers the interaction from text mining, and a blue line indicates the predictive function relationship. By holding the cursor over each protein, the related annotation for the protein is displayed and allows the user to navigate the network and easily check a proteins’ relationship. ‘Text Format Output’ will export the interaction pair information in text format.
Figure 4.
AtPID interface of the (Network Display) window for the queried protein, HAP3. In the (Network Display) window, we can view the interaction network involved with queried protein(s) and the neighbor component(s) intuitively. (Text Format Output) can export the interaction pair information for further analysis.
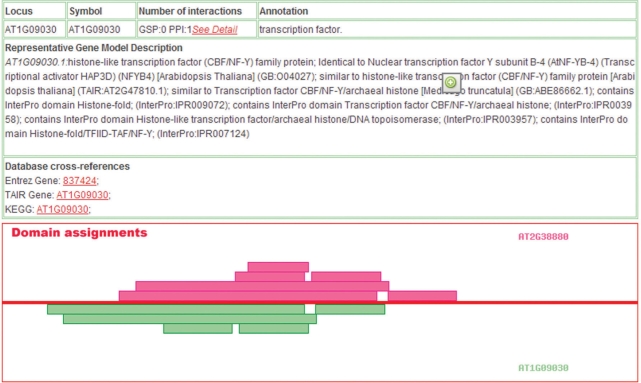
In the ‘Pair Search Results’ window when users submit potential interaction pairs, the domain attribute of one partner protein (e.g. AT1G09030) can be viewed graphically (Figure 5). Each module is linked to Pfam and when the user places the cursor over the module, details of the domain will also be displayed. Thus, AtPID provides comprehensive knowledge through a friendly and convenient interface that should be easy to use by biologists.
Figure 5.
Domain attributes of one interactant of the queried protein, HAP3A.
Software development
The database server is located at SIBS (Shanghai Institute of Biological Science) data service platform. Therefore, clients around the world can readily access the AtPID database. The AtPID development environment is apache+php+mysql, which allows for more efficient calculation rate performances and augmentation of the program.
CONCLUSIONS
AtPID is an online repository of A. thaliana protein interactions. AtPID serves as a major reference site for PPIs using Arabidopsis as a model plant system. The database collection will regularly integrate new accessions as they become available. A number of new features and applications are currently under construction, such as a gene regulation and dynamic PPI network that function under different conditions with increased gene expression and proteomics data.
Currently, the subcellular localization predictions of A. thaliana are available for both the chloroplast and mitochondrion and the predictive organellar proteins have been added into AtPID. In addition, we plan to conduct further assessments of proteins to other cellular and/or subcellular locations, including nuclear, cytoplasmic and extracellular proteins.
Another important field of research is to elucidate the relationships between phenotype and genotype. For example, we plan to collect data relevant to mutants and their respective phenotypes. These highly varied types of data will be available through AtPID in the near future.
ACKNOWLEDGEMENTS
This work was supported by the State Key Program of Basic Research of China grants (2002CB713807, 2007CB108800), the National High Technology Research and Development Program of China (863 project) (Grant No. 2006AA02Z313, 2006AA10Z129) and National Natural Science Foundation of China grants (90408010 and 30571510). Funding to pay the Open Access publication charges for this article was provided by National Natural Science Foundation of China and the State Key Program of Basic Research of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Magdalena S. Network biology: a protein network of one's own proteins. Nat. Rev. Genets. 2005;6:800. [Google Scholar]
- 2.Uhrig JF. Planta. 2006:1–11. doi: 10.1007/s00425-006-0260-x. [DOI] [PubMed] [Google Scholar]
- 3.Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nat. Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Zhao H. A computational approach for ordering signal transduction pathway components from genomics and proteomics Data. BMC Bioinformatics. 2004;5:158–163. doi: 10.1186/1471-2105-5-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol. Syst. Biol. 2005;0008 doi: 10.1038/msb4100012. msb4100012:E4100011–E410001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakakibara H, Takei K, Hirose N. Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci. 2006;11:440–448. doi: 10.1016/j.tplants.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Reed JL, Famili I, Thiele I, Palsson BO. Towards multidimensional genome annotation. Nat. Rev. Genet. 2006;7:130–141. doi: 10.1038/nrg1769. [DOI] [PubMed] [Google Scholar]
- 8.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 10.Giot L, Bader J, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Armstrong C, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 13.Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29:102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Folter S, Immink R, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hackbusch J, Richter K, Müller J, Salamini F, Uhrig JF. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl Acad. Sci. USA. 2005;102:4908–4912. doi: 10.1073/pnas.0501181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Formosa T, Barry J, Alberts BM, Greenblatt J. Using protein affinity chromatography to probe structure of protein machines. Meth. Enzymol. 1991;208:24–45. doi: 10.1016/0076-6879(91)08005-3. [DOI] [PubMed] [Google Scholar]
- 17.Miller KA, Alberts BM. F-actin affinity chromatography: technique for isolating previously unidentified actin-binding proteins. Proc. Natl Acad. Sci. USA. 1989;86:4808–4812. doi: 10.1073/pnas.86.13.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller KG, Field CM, Alberts BM. Use of actin filament and microtubule affinity chromatography to identify proteins that bind to the cytoskeleton. Meth. Enzymol. 1991;196:303–319. doi: 10.1016/0076-6879(91)96028-p. [DOI] [PubMed] [Google Scholar]
- 19.Baird BA, Hammes GG. Chemical cross-linking studies of chloroplast coupling factor 1. J. Biol. Chem. 1976;251:6953–6962. [PubMed] [Google Scholar]
- 20.Bragg P, Hou C. A cross-linking study of the Ca2+, Mg2+-activated adenosine triphosphatase of Escherichia coli. Eur. J. Biochem. 1980;106:495–503. doi: 10.1111/j.1432-1033.1980.tb04596.x. [DOI] [PubMed] [Google Scholar]
- 21.Cover J, Lambert JM, Norman CM. Identification of proteins at the subunit interface of the Escherichia coli ribosome by cross-linking with dimethyl 3,3'-dithiobis(propionimidate) Biochemistry. 1981;12:2843–2852. doi: 10.1021/bi00513a021. [DOI] [PubMed] [Google Scholar]
- 22.Krieg U, Johnson AE, Walter P. Protein translocation across the endoplasmic reticulum membrane: identification by photocross-linking of a 39-kD integral membrane glycoprotein as part of a putative translocation tunnel. J. Cell Biol. 1989;109:2033–2043. doi: 10.1083/jcb.109.5.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 24.Parrish JR, Gulyas K, Finley RL. Yeast two-hybrid contributions to interactome mapping. Curr. Opin. Biotechnol. 2006;17:387–393. doi: 10.1016/j.copbio.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Chien C, Bartel PL, Sternglanz R. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl Acad. Sci. USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clackson T, Hoogenboom HR, Griffiths AD. Making antibody fragments using phage display libraries. Nature. 1991;352:624–628. doi: 10.1038/352624a0. [DOI] [PubMed] [Google Scholar]
- 27.Phizicky EM, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol. Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deane CM, Salwinski Ł., Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell. Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 29.Fu LD, Tsamardinos I. AMIA Annu. Symp. Proc. 2005. A comparison of Bayesian network learning algorithms from continuous data; p. 960. [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Li X, Su H, Chen H, Galbraith DW. A framework of integrating gene relations from heterogeneous data sources: an experiment on Arabidopsis thaliana. Bioinformatics. 2006;22:2037–2043. doi: 10.1093/bioinformatics/btl345. [DOI] [PubMed] [Google Scholar]
- 31.Needham CJ, Bradford JR, Bulpitt AJ, Westhead DR. Inference in Bayesian networks. Nat. Biotechnol. 2006;24:51–54. doi: 10.1038/nbt0106-51. [DOI] [PubMed] [Google Scholar]
- 32.Kato T, Tsuda K, Asai K. Selective integration of multiple biological data for supervised network inference. Bioinformatics. 2005;21:2488–2495. doi: 10.1093/bioinformatics/bti339. [DOI] [PubMed] [Google Scholar]
- 33.Daeui P, Semin L, Dan B, Michael S, Michael L, Donghoon O, Jong B. Comparative interactomics analysis of protein family interaction networks using PSIMAP (protein structural interactome map) Bioinformatics. 2005;21:3234–3240. doi: 10.1093/bioinformatics/bti512. [DOI] [PubMed] [Google Scholar]
- 34.Walhout AJ, Vidal M. Protein interaction maps for model organisms. Nat. Rev. Mol. Cell Biol. 2001;2:55–62. doi: 10.1038/35048107. [DOI] [PubMed] [Google Scholar]
- 35.Ben-Hur A, Noble WS. Kernel methods for predicting protein-protein interactions. Bioinformatics. 2005;21(Suppl. 1):i38–i46. doi: 10.1093/bioinformatics/bti1016. [DOI] [PubMed] [Google Scholar]
- 36.Buckingham SD. Data mining for protein-protein interactions in invertebrate model organisms. Invert. Neurosci. 2005;5:183–187. doi: 10.1007/s10158-005-0009-4. [DOI] [PubMed] [Google Scholar]
- 37.Zhong W, Sternberg PW. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311:1381–1382. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
- 38.Moon HS, Bhak J, Lee KH, Lee D. Architecture of basic building blocks in protein and domain structural interaction networks. Bioinformatics. 2005;21:1479–1486. doi: 10.1093/bioinformatics/bti240. [DOI] [PubMed] [Google Scholar]
- 39.Patil A, Nakamura H. Filtering high-throughput protein-protein interaction data using a combination of genomic features. BMC Bioinformatics. 2005;18:100. doi: 10.1186/1471-2105-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhodes DR, Tomlins SA, Varambally S, Mahavisno V, Barrette T, Kalyana-Sundaram S, Ghosh D, Pandey A, Chinnaiyan AM. Probabilistic model of the human protein-protein interaction network. Nat. Biotechnol. 2005;23:951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- 41.Valente AX, Cusick ME. Yeast protein interactome topology provides framework for coordinated-functionality. Nucleic Acids Res. 2006;34:2812–2819. doi: 10.1093/nar/gkl325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Date SV, Stoeckert CJ. Computational modeling of the Plasmodium falciparum interactome reveals protein function on a genome-wide scale. Genome Res. 2006;16:542–549. doi: 10.1101/gr.4573206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suthram S, Sittler T, Ideker T. The Plasmodium protein network diverges from those of other eukaryotes. Nature. 2005;438:108–112. doi: 10.1038/nature04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, et al. IntAct – open source resource for molecular interaction data. Nucleic Acids Res. 2007;35(Database issue):D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paul JK, Jorge D, Allyson W, Youla K, Ewan B, Rolf A. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 47.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34(Database issue):357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matthews LR, Vaglio P, Reboul J, Ge H, Davis BP, Garrels J, Vincent S, Vidal M. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Genome Res. 2001;11:2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ge H, Liu Z, Church GM, Vidal M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet. 2001;29:482–486. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- 50.Grigoriev A. A relationship between gene expression and protein interactions on the proteome scale: analysis of the bacteriophage T7 and the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:3513–3519. doi: 10.1093/nar/29.17.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 52.Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 54.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 55.Huynen M, Snel B, Lathe W., III, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pellegrini M, Marcotte E, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun J, Xu J, Liu Z, Liu Q, Zhao A, Shi T, Li Y. Refined phylogenetic profiles method for predicting protein-protein interactions. Bioinformatics. 2005;21:3409–3415. doi: 10.1093/bioinformatics/bti532. [DOI] [PubMed] [Google Scholar]
- 58.O'Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33(Database issue):D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salwinski L, Miller C, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32(Database issue):D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 61.Heazlewood JL, Verboom R, Tonti-Filippini J, Small I, Millar AH. SUBA: the Arabidopsis subcellular database. Nucleic Acids Res. 2007;35:D213–D218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hooper SD, Bork P. Medusa: a simple tool for interaction graph analysis. Bioinformatics. 2005;21:4432–4433. doi: 10.1093/bioinformatics/bti696. [DOI] [PubMed] [Google Scholar]
- 63.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 65.Small I, Peeters N, Legeai F, Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- 66.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]