Abstract
ARED Organism represents the expansion of the adenylate uridylate (AU)-rich element (ARE)-containing human mRNA database into the transcriptomes of mouse and rat. As a result, we performed quantitative assessment of ARE conservation in human, mouse and rat transcripts. We found that a significant proportion (∼25%) of human genes differ in their ARE patterns from mouse and rat transcripts. ARED-Integrated, another updated and expanded version of ARED, is a compilation of ARED versions 1.0 to 3.0 and updated version 4.0 that is devoted to human mRNAs. Thus, ARED-Integrated and ARED-Organism databases, both publicly available at http://brp.kfshrc.edu.sa/ARED, offer scientists a comprehensive view of AREs in the human transcriptome and the ability to study the comparative genomics of AREs in model organisms. This ultimately will help in inferring the biological consequences of ARE variation in these key animal models as opposed to humans, particularly, in relationships to the role of RNA stability in disease.
INTRODUCTION
The adenylate uridylate (AU)-rich elements (AREs) that commonly exist in the 3′-untranslated regions (3′UTR) constitute a major mRNA destabilization determinant. More specifically, the pentamer AUUUA existing in U-rich context has been shown to be a discriminating motif for the AREs that generally consist of tandem overlapping repeats of this pentamer (1,2). The nonamer UUAUUUAUU has been considered a minimal functional ARE motif (3). In earlier work, we have shown that the patterns WWUUAUUUAUUWW and WWWUAUUUAUUUW are highly specific to the 3′UTR of the mRNA compared with the 5′UTR and the coding regions of the transcriptome and are more functionally related to mRNA stability (4,5).
Functional AREs have been characterized in a number of gene products that include critical players in cellular growth and innate immunity and were linked to certain disease states (6–8). Although ARE-mRNAs were found to code for a functionally diverse group of proteins, they are overrepresented in specific functional categories such as cell proliferation, RNA metabolism, transcriptional regulation, signaling, response to stress and microbes and developmental processes (5). Thus, the importance of this class of genes and their relationship with disease motivated the establishment of the ARE database to identify the repertoire of genes that are potentially regulated by ARE-mediated mRNA turnover by using bioinformatics. Earlier, we focused on the identification of ARE motifs and the development of algorithms that led to the identification and cataloguing of ARE-mRNAs in the human genome. Furthermore, we have updated ARED (ARED 4.0) and integrated the three previous databases into one Integrated ARED. Due to their prominent role as model experimental systems, the genomics of laboratory mice is second in importance to human genomics. This has led us to expand ARED to mouse and rat in addition to updating the collection of human ARE-mRNAs. The database should be valuable to researchers in many different fields since ARE genes are functionally heterogeneous (5) and ARE variation in the mouse/rat transcriptome could lead to important inferences in mRNA biology between organisms. Here we present ARED Organism, an extension of ARED to non-human organisms, and ARED-Integrated, a compilation of earlier releases with an updated data.
MATERIALS AND METHODS
Data sources
ENSEMBL (9) data were downloaded from BioMart (http://www.biomart.org). We used Human ENSEMBL release 44, Mouse ENSEMBL release 42 and Rat ENSEMBL release 44. The 3′UTR regions in ENSEMBL were computationally extracted from the corresponding genomic builds that are, respectively, NCBI build 36, NCBI build m36 and RGSC 3.4. Cross references to RefSeq, UNIGENE, Entrez and the gene ontology (GO) terms were also downloaded from BioMart for the same database releases. We used the BioMart homology database, based on ENSEMBL release 46 for human, mouse and rat organisms. These datasets were used for ARED Organism. ARED 4.0 was generated from RefSeq release 16. ARED 4.0 was cross-referenced with Unigene (release 190), Gene Ontology, GO (through Entrez, release dated 20 April 2006), as well as the GenBank definition lines (release 153). The three AREDs were merged into one database along with Version 4.0, following the structure of the latest ARED 3.0 release (http://brp.kfshrc.edu.sa/ARED/), and called ARED Integrated. Algorithms used in this build up were previously described (5).
ARE identification and database construction
The 3′UTR sequence data were retrieved as FASTA files using Biomart sequence options. We then scanned for the presence of the polyA signal (AWTAAA) in the last 50 bases of each sequence. We used ‘scan_for_matches’ [SFM, (10)] to analyze the 3′UTR sequence data and search for ARE motifs. SFM is a publicly available tool that allows searching sequence data for a pattern, allowing mismatching and including insertion and deletion. The patterns of AREs were described in the previous versions of ARED papers (5,11,12). In the database, the transcripts were marked for the presence of a complete 3′UTR and a polyA signal. Results were cross-linked to gene definitions and other database identifiers such as Unigene, RefSeq and GO. Gene definitions were keyword indexed and made searchable so that a user may also query ARED using keywords.
ARED entries are based on genes, but the analyses were carried out on the transcripts themselves. A gene is assigned based on the most stringent ARE class among its transcripts, whereas ‘Non-ARE-containing’ is assigned to a gene if none of its transcripts contains an ARE. Thus, a gene is ARE-containing if it is protein coding and has the minimal ARE motif in its 3′UTR.
ARE gene ortholog dataset compilation
The homology data were cross-matched to the ARE pattern cluster using the following criteria: (i) each gene has one ortholog in the other organism, (ii) both genes in a pair must be known genes, (iii) the transcripts used as evidence in our ARE cluster classification must be the same transcripts used to infer homology in both organisms, (iv) pairs were excluded if both genes do not contain an ARE and (v) pairs were excluded if AREs are absent and 3′UTRs are incomplete.
Orthology statistical analysis
As a result of this extraction approach, a list was created for each organism pair containing the matched gene pairs and the assigned ARE cluster groups (c1, c2 variables). We then assigned non-ARE genes a score of 0 and the rest of the genes scores corresponding to the count of AUUUA pentamers in the ARE cluster group (i.e. Cluster Group I is assigned a score of 5 and Group II assigned 4, whereas Group IV—the weakest—is assigned a score of 1). The count of pentamers in any cluster is the number of overlapping continuous repeats of the AUUUA pentamer. The different ARE clusters were previously described (11). Genes with multiple AREs are assigned based on the highest counts of AUUUA pentamers, i.e. the longest stretch of ARE. For the Chi-squared test, c1 and c2 were treated as two random variables; the 6 × 6 (values 0–5 for each variable) contingency table was populated with the number of times each pair was observed in the list. Expected values were then calculated and the Chi-squared statistic is obtained. We observed that some expected values (especially for cells where both c1 and c2 were 4 or 5) were small (<5) so we repeated the computation after collapsing values 4 and 5 onto 3. The results remained highly significant for all organism pair tables tested.
For the difference distribution test, we calculated the difference c1−c2 and measured the observed frequency of each difference value (−5 to +5, or 0–5 for the unsigned test). We carried out an empirical shuffling experiment to generate a random background distribution. Repeated runs of the randomization experiments produced consistent results showing that the observed distribution had higher information content than the random background. Absolute difference measurements also showed the same result.
RESULTS AND DISCUSSION
Two main schemes were adopted to comprehensively compile human ARE-mRNAs and ARE-genes and to facilitate comparative genomics of AREs among human, mouse and rat species. ARED-Integrated is an expanded and update human ARE-mRNA/gene database that is built by combining ARED versions 1.0, 2.0 and 3.0 and an updated version (ARED 4.0) in order to compile the different algorithms and search strategies used for ARE mining. ARED-Integrated contained 3700 ARE-genes and 6153 RefSeq transcripts. This translates to ∼10% of total genes.
The other important development is ARED Organism, which in addition to human data included a compilation of AREs and ARE-mRNAs from the widely analyzed genomes of mouse and rat. We chose these two organisms due to their importance as laboratory in vivo models and the availability of large proportion of transcript sequences. Future expansions of the collection to include other organisms will only require the availability of a reasonably well-cataloged transcriptome rich in complete 3′UTRs. According to ARED Organism that is based on ENSEMBL, at least 7% of human genes are ARE genes. Since 41.5% of human ENSEMBL genes lack 3′UTR sequences (18 452 genes), ARE genes constitute ∼11% of the total gene number in human ENSEMBL. While at least 5% of mouse and 2% of rat genes contain ARE motif, these smaller percentages are due to the higher proportion of genes without annotated 3′UTRs, when compared with human genes.
ARED Organism allowed us to study ARE cluster variations among the organism pairs. Based on strict criteria, we extracted 907 pairs of orthologous human/mouse genes from 23 035 pairs, 394 from 20 715 human/rat pairs and 411 from 26 695 mouse/rat pairs. Due to subtle deviations in ARE clusters, for example, due to non-functional mutations or sequencing errors, numerical score assignment (0–5, depending on the number of pentamers) may deviate slightly from the ‘true’ value. We therefore compared the classes of the gene pairs by examining the distribution of both the signed and absolute difference in the ARE scores between matched gene pairs (Figure 1). We generated randomized pairings by shuffling (a thousand times) the cluster score within each ortholog list and obtained corresponding distributions. We finally compared the information content in the observed versus randomized distributions and found it to be significantly larger in all cases (P < 0.005). Using conventional Chi-squared test, we found that the probability of independence is extremely unlikely (−log P > 30) for all three organism pairs. Thus, both methods support statistical significance of overall ARE conservation among the three organisms.
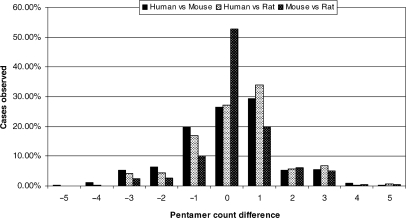
Figure 1.
AUUUA pentamer count-difference distribution among ARE genes in the orthologs. X-axis denotes score differences in pentamers between orthologs in each organism pair and Y-axis denotes the observed frequency of each difference.
Although the analysis shows that many of the ARE clusters are highly conserved between the pairs of human, mouse and rat species, there is an appreciable number of orthologs that have significant ARE cluster variations. There were ∼25%, and 21% of ARE orthologs that have significant differences—AREs differed by at least two cluster scores—in human/mouse and human/rat. If those with weaker variations (i.e. with at least one cluster score) would be included, only less than half of the ARE clusters are completely conserved between human and mouse or rat. These variations comprise either absence or presence of an ARE or the number of the overlapping pentamer repeats (Table 1). Certain human genes may code for unstable ARE-mRNAs but their mouse and/or rat homologs code for potentially stable non-ARE mRNA. For example, in humans, the FUT1 gene that codes galactoside 2-alpha-l-fucosyltransferase 1 mRNA has an ARE Cluster I (i.e. five pentamer stretch), whereas the mouse and rat FUT1 mRNAs lack an ARE. This may result in significant difference in the biology of cancer maintenance between human and mouse since it has been found that stabilization of FUT1 ARE-mRNA in human cancer cells leads to enhanced oxygen-dependent glycolysis in the cancer cells (13). The PIM-1 proto-oncogene codes for an unstable mRNA Cluster I ARE in humans while in rat, the PIM-1 mRNA lacks any ARE. The NRAMP and CD69 that code for proteins important in immunity harbor a Cluster III ARE in human mRNAs but lack in ARE in the mouse mRNAs. It is also possible that mouse or rat mRNA contains functional AREs when compared with human mRNAs. For example, the Cluster I ARE in the mouse nuclear FMRP-interacting protein 1 mRNA that codes for nuclear fragile X mental retardation protein interacting protein 2 is absent in the human mRNA. Another notable mouse mRNA is ICAM1 that has three pentamer repeats yet no ARE is found in the human mRNA. Although mouse and rat are closely related species, ∼17% of their ARE clusters differ; examples with large ARE cluster variations are dual-specificity phosphatase 11 (DUSP11) and Protein-arginine deiminase type-2 (PADI2). Complete list of the ARE orthologs are found in the ARED website (http://brp.kfshrc.edu.sa/ared). It should be noted that ARE orthology analysis has limitations inherited from the originally obtained homology data, and, as this is a bioinformatics article, true assessment of functional AREs variations in orthologs requires experimental validation.
Table 1.
ARE orthologs with ARE variations: examples of those with three or more ARE score differencea
| Gene | Definition | Human | Mouse | Rat |
|---|---|---|---|---|
| FUT1 | fucosyltransferase 1 | 5 | 0 | 0 |
| PIM1 | Proto-oncogene serine/threonine-protein kinase Pim-1 | 5 | 0 | 0 |
| DUSP11 | Dual-specificity protein phosphatase 11 | 0 | 5 | na |
| NUFIP1 | Nuclear FMRP-interacting protein 1 | 1 | 5 | 0 |
| SLC11A1 | Natural resistance-associated macrophage protein 1 (NRAMP 1) | 4 | 0 | na |
| BET1 | Golgi vesicular membrane-trafficking protein (p18) | 4 | 0 | 1 |
| TMEM53 | Transmembrane protein 53 | 4 | 0 | na |
| KIF1C | Kinesin-like protein KIF1C | 4 | 0 | na |
| KATNAL1 | Katanin p60 subunit A-like 1 | 4 | 0 | 0 |
| OSMR | Oncostatin M receptor | 4 | 0 | na |
| SLC25A32 | Mitochondrial folate transporter/carrier | 4 | 0 | na |
| PNOC | Nociceptin precursor | 0 | 4 | 0 |
| SLC1A7 | Excitatory amino acid transporter 5 | 0 | 4 | na |
| XKR8 | XK-related protein 8 | 0 | 4 | na |
| EVA1 | Epithelial V-like antigen 1 precursor | 0 | 4 | 4 |
| ETV6 | ETS variant gene 6 (TEL oncogene) | 0 | 4 | na |
| PPP1R15A | Protein phosphatase 1, regulatory subunit 15A | 1 | 5 | 3 |
| PLEKHA8 | Pleckstrin homology domain containing, family A member 8 | 0 | 4 | na |
| PADI2 | Peptidyl arginine deiminase, type II | na | 4 | 0 |
| UTB18 | UTP18, small subunit (SSU) processome component | na | 0 | 3 |
| TNFRSF 11B | Tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) | 0 | 0 | 3 |
| TRPM2 | Short transient receptor potential channel 1 | na | 0 | 3 |
| THEM4 | Thioesterase superfamily member 4 | 0 | 0 | 3 |
| SLC17A6 | Solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 | na | 0 | 3 |
| TRPS1 | Zinc finger transcription factor Trps1 | 3 | 0 | na |
| ADAM2 | A disintegrin and metallopeptidase domain 2 | 3 | 0 | na |
| CD69 | Early activation antigen CD69 | 3 | 0 | 0 |
| TANK | TRAF family member-associated Nf-kappa B activator | 3 | 0 | na |
| ICAM1 | Intercellular adhesion molecule 1 precursor (ICAM-1) | 1 | 3 | na |
| BIRC3 | Inhibitor of apoptosis protein 1 | 3 | 0 | na |
| CBLN3 | Cerebellin 3 precursor protein | 0 | 3 | na |
| FOXP1 | Forkhead box protein P1 | 3 | na | 0 |
| FGL2 | Fibrinogen-like protein 2 | 0 | 1 | 3 |
| LEO1 | RNA polymerase-associated protein LEO1 | 0 | 1 | 3 |
| PRKX | Protein kinase, X-linked | na | 0 | 3 |
aNumbers shown are cluster types. Complete listing is a part of ARED Organism, http://brp.kfshrc.edu.sa/ared. na: not available.
Despite the fact that many well-studied ARE genes such as TNF-α, IL-1 and c-fos have well-conserved ARE patterns, the analysis reveals that many other genes, although not studied for their mRNA stability, have significant differences in their ARE patterns. The data here may suggest that ARE-mediated post-transcriptional control for a given gene may be functional in one organism but not in the other despite being orthologs. This may underlie significant biological ramifications between human and mouse biology in relationship to RNA stability in disease development and immune response.
ACKNOWLEDGEMENTS
The work was supported by King Abdelaziz City for Science and Technology (KACST; grant AP-24-56 to K.S.A.K). We thank Dr Edward Hitti for reading the manuscript. Funding to pay the Open Access publication charges for this article was provided by KACST.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 2.Chen CY, Shyu AB. Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol. Cell. Biol. 1994;14:8471–8482. doi: 10.1128/mcb.14.12.8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zubiaga AM, Belasco JG, Greenberg ME. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raghavan A, Dhalla M, Bakheet T, Ogilvie RL, Vlasova IA, Khabar KS, Williams BR, Bohjanen PR. Patterns of coordinate down-regulation of ARE-containing transcripts following immune cell activation. Genomics. 2004;84:1002–1013. doi: 10.1016/j.ygeno.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khabar KS, Young HA. Post-transcriptional control of the interferon system. Biochimie. 2007;89:761–769. doi: 10.1016/j.biochi.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khabar KS. Rapid transit in the immune cells: the role of mRNA turnover regulation. J. Leukoc. Biol. 2007;81:1335–1344. doi: 10.1189/jlb.0207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eberhardt W, Doller A, Akool el S, Pfeilschifter J. Modulation of mRNA stability as a novel therapeutic approach. Pharmacol. Ther. 2007;114:56–73. doi: 10.1016/j.pharmthera.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- 11.Bakheet T, Frevel M, Williams B.RG, Greer W, Khabar K.SA. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional reportiore of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bakheet T, Williams BR, Khabar KS. ARED 2.0: an update of AU-rich element mRNA database. Nucleic Acids Res. 2003;31:421–423. doi: 10.1093/nar/gkg023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chesney J, Mitchell R, Benigni F, Bacher M, Spiegel L, Al-Abed Y, Han JH, Metz C, Bucala R. An inducible gene product for 6-phosphofructo-2-kinase with an AU-rich instability element: role in tumor cell glycolysis and the Warburg effect. Proc. Natl Acad. Sci. USA. 1999;96:3047–3052. doi: 10.1073/pnas.96.6.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]