Abstract
Interleukin 12 (IL-12)-induced T helper 1 (Th1) development requires Stat4 activation. However, antigen-activated Th1 cells can produce interferon γ (IFN-γ) independently of IL-12 and Stat4 activation. Thus, in differentiated Th1 cells, factors regulated by IL-12 and Stat4 may be involved in IFN-γ production. Using subtractive cloning, we identified ERM, an Ets transcription factor, to be a Th1-specific, IL-12-induced gene. IL-12-induction of ERM occurred in wild-type and Stat1-deficient, but not Stat4-deficient, T cells, suggesting ERM is Stat4-inducible. Retroviral expression of ERM did not restore IFN-γ production in Stat4-deficient T cells, but augmented IFN-γ expression in Stat4-heterozygous T cells. Ets factors frequently regulate transcription via cooperative interactions with other transcription factors, and ERM has been reported to cooperate with c-Jun. However, in the absence of other transcription factors, ERM augmented expression of an IFN-γ reporter by only 2-fold. Thus, determining the requirement for ERM in Th1 development likely will require gene targeting.
Protection from pathogens requires appropriate effector responses, which can be enhanced by emergence either T helper (Th)1 or Th2 phenotype CD4+ T cells (1, 2). For example, Th1 development characterized by production of interferon γ (IFN-γ), but not interleukin 4 (IL-4), aids in elimination of the intracellular pathogens Listeria monocytogenes and Leishmania major (1). Th1 development in CD4+ T cells requires IL-12, which is produced by activated macrophages (3) or dendritic cells (4, 5) and which, in murine T cells, uniquely activates the transcription factor Stat4 (6, 7). The importance of Stat4 in IL-12-induced Th1 development was demonstrated by the diminished IFN-γ response to IL-12 by Stat4-deficient T cells (8, 9). Similarly, IL-4-induced Th2 development requires activation of Stat6 (10–12).
Two transcription factors, c-Maf and GATA3, have been found to be expressed selectively by Th2 cells (13, 14). These factors influence IL-4 production by direct promoter interactions (13) or by effects on the IL-4 locus (15, 16). In contrast, no Th1-specific transcription factors have been identified that clearly regulate IFN-γ transcription. Whereas Stat4 is expressed by Th1 and Th2 cells (17), Stat4 activation by IL-12 occurs only in Th1 cells because of Th1-restricted expression of the IL-12R β2 subunit (18). One study suggested that Stat4 may participate directly in IFN-γ regulation by binding nonconsensus, low-affinity STAT sites in the IFN-γ promoter and first intron (19), but did not examine the role of these sites in native IFN-γ regulation. In this model, Stat4 would directly augment IFN-γ gene expression in Th1 cells either alone or in cooperation with other transcription factors. In either case, activated Stat4 would be required for cytokine expression in fully differentiated Th1 cells.
Here we show that T cell antigen receptor (TCR)-induced IFN-γ transcription in differentiated Th1 cells does not require Stat4 activation. We identify an Ets family transcription factor, ERM, as an IL-12-induced, Stat4-dependent gene selectively expressed by Th1 cells. However, ERM expression does not restore IFN-γ production in Stat4-deficient T cells, suggesting that either it regulates some other aspect of Th1 behavior besides IFN-γ production or that ERM requires cooperation with other Th1-specific factors in regulating IFN-γ expression.
METHODS
T Cell Purification and Activation.
Stat1- and Stat4-targeted heterozygous founder mice, gifts of R. D. Schreiber (20) and J. N. Ihle (9), were backcrossed three times to DO11.10 TCR-transgenic BALB/c mice before interbreeding to generate Stat1−/− and Stat4−/− H-2d DO11.10 TCR-transgenic animals. Splenocytes from DO11.10 or BALB/c mice were purified on a density gradient (Histopaque-1119, Sigma) and activated by 0.3 μM of ovalbumin peptide 323–339 (OVA) (3) or 1 μg/ml of Con A (Sigma), respectively. For anti-CD3 stimulation, plates were coated with 5 μg/ml of 500A2 (gift of R. D. Schreiber) overnight at 4°C. Transgenic T cells (1.25 × 105/ml) were reactivated with 0.3 μM of OVA peptide by using irradiated BALB/c splenocytes (2.5 × 106/ml) for antigen-presenting cells (APCs) as described (21). On day 7, T cells were washed, counted, and restimulated as indicated. Supernatants or RNA was harvested at 48 hr of stimulation or as indicated. The Th1 clone 3F6, described previously (6), was maintained by biweekly antigen stimulation. Media, cytokines, and antibodies were as described previously (21, 22).
RNA, Northern Blots, and cDNA Library.
Total RNA was purified by RNeasy system (Qiagen, Chatsworth, CA). Poly(A) RNA was isolated by PolyA-Tract mRNA Isolation System IV (Promega). Double-stranded cDNA and cDNA library was synthesized by cDNA synthesis kit and ZAP-cDNA Gigapack II gold cloning system (Stratagene). For Northern blots, 10 μg total RNA was electrophoresed and transferred to Zeta Probe membrane (Bio-Rad). For construction of cDNA libraries, BALA/c splenocytes were activated by Con A with either IL-12 or IL-4 to initiate Th1 or Th2 development, respectively, and RNA was isolated after 2 days. The Th1 library was screened by using RDA product D15 (described below) as probe. Greater than 2 × 106 independent plaques were screened to obtain two separate, full-length murine ERM cDNA isolates.
Immunoprecipitation and Western Blot Analysis.
Stat4 was immunoprecipitated with NB34 mAb as described (6, 23). Proteins were separated by SDS/PAGE, probed with antiphosphotyrosine RC20 (1:7,500; Transduction Laboratories, Lexington, KY), and then stripped and probed with NB34 (1 μg/ml).
Representational Difference Analysis (RDA).
Splenocytes from BALB/c mice were stimulated by Con A plus either IL-12 or IL-4 to initiate Th1 or Th2 development. RDA was performed as described (24) with minor modifications. Double-stranded cDNA from day 2 Th1 and Th2 cultures was digested by Tsp509I or DpnII (New England Biolabs), and RDA was done separately for each digest. The sequences of R-12-Tsp509I, J-12-Tsp509I, and N-12-Tsp509I are 5′-AATTTGCGGTGA-3′, 5′-AATTTGTTCATG-3′, and 5′-AATTTTCCCTCG-3′, respectively. The remaining oligonucleotides are as described (24). Three rounds of hybridization subtraction were performed using by Tester (Th1)/Driver (Th2) ratios of 1:100, 1:800, and 1:40,000, sequentially. Final PCR products were digested with either Tsp509I or DpnII, ligated into pBSSK vector at EcoRI (for Tsp509I) or BamHI (for DpnII) sites, and transformed into DH5α-competent cells, and individual colonies were isolated and RDA inserts were recovered by PCR using T3 and T7 primers. PCR products were dot-blotted onto duplicate filters and hybridized with probes from Tester and Driver representations. Colonies with selectively greater hybridization to Tester compared with Driver were used as probes in Northern hybridization against Th1 and Th2 RNA. Two hundred individual colonies were screened, and 41 were equivalent to one designated D15.
Retroviral Constructs and Retroviral Transduction of Naïve T Cells.
The vector GFP-RV has been described (15). Phoenix-Eco packaging cells were from G. Nolan (Stanford University, Stanford, CA). Viral supernatant was produced according to Nolan’s protocol (http://www.stanford.edu/group/nolan/NL-phnxr.html) except that T cell infection was augmented by using RetroNectin (Takara Shuzo, Kyoto). Two-day activated T cells were cultured with retroviral supernatant on RetroNectin-coated plates for 2 days and transferred to fresh plates and medium for 3 more days. T cells were sorted for GFP expression on day 7.
IFN-γ Promoter Reporter Analysis.
IFN-γ-promoter reporter constructs F and H have been described previously (16). The full-length ERM cDNA was cloned into the BamHI and XhoI sites of pcDNA3.1 expression vector (Invitrogen) to produce pcDNAERM. Jurkat cells (107) were mixed with 20 μg IFN-γ promoter reporter construct, 10 μg pcDNA or pcDNAERM, and 1 μg CMV-Renilla luciferase plasmid (15) in 1.2 ml RPMI 1640 medium. Cells were electroporated in three 400-μl aliquots at 960 μF and 280 V by using a Bio-Rad Gene Pulser, combined, and supplied with 1 ml fresh media. After 12–14 hr, cells were divided into two parts and one was stimulated with 50 ng/ml of PMA and 1 μM of ionomycin for 4 hr before luciferase assay (25).
RESULTS
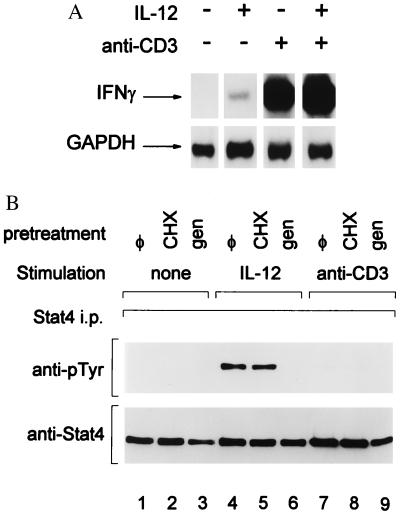
We first asked whether IFN-γ production by a differentiated Th1 clone 3F6 required concurrent Stat4 activation (Fig. 1). Treatment of 3F6 cells with anti-CD3, either with or without added IL-12, led to marked induction of IFN-γ mRNA (Fig. 1A, lanes 3 and 4). In contrast, treatment with only IL-12 led to nearly undetectable IFN-γ induction (Fig. 1A, lane 2). To ask whether the absence of IL-12-induced IFN-γ was simply due to an absence of IL-12 receptors on 3F6 cells, we examined IL-12-induced Stat4 activation (Fig. 1B). 3F6 cells clearly express IL-12 receptors because treatment with IL-12 induced specific tyrosine phosphorylation of Stat4 (Fig. 1B). This phosphorylation occurred both in the presence and absence cycloheximide (Fig. 1B, lanes 4 and 5), but not in the presence of a tyrosine kinase inhibitor, Genistein (lane 6), as expected (6). In contrast, anti-CD3-treated 3F6 cells did not activate Stat4 (lanes 7–9). Thus, anti-CD3-induced IFN-γ production in differentiated Th1 cells is independent of Stat4 activation, and IL-12-induced Stat4 activation alone is insufficient for IFN-γ production.
Figure 1.
IFN-γ production induced by anti-CD3 stimulation in differentiated Th1 cells is IL-12- and Stat4-indepenent. (A) 3F6 T cells were stimulated (+) with anti-CD3 or IL-12 (10 units/ml) as indicated or left unstimulated (−) for 4 hr as described in Methods. Total RNA was prepared and Northern blot analysis for IFN-γ was performed (17). The autoradiogram was overexposed to allow detection of the faint band present in the IL-12-treated lane. The blot was stripped and reprobed for GAPDH as a loading control as described (17). (B) 3F6 T cells were pretreated with Genistein (50 μg/ml) (gen) or cycloheximide (10 μg/ml) (CHX) or left untreated (φ), as indicated, for 30 min and then stimulated with IL-12 (10 units/ml) or anti-CD3 for 30 min, or untreated (none) as indicated. Cells were lysed, and immunoprecipitation for Stat4 was performed by using NB34 as described (17). Western detection of phosphotyrosine was performed with antiphosphotyrosine RC20 (Transduction Laboratories), and blots were stripped and reprobed with anti-Stat4 (Lower) to indicate uniform Stat4 immunoprecipitation.
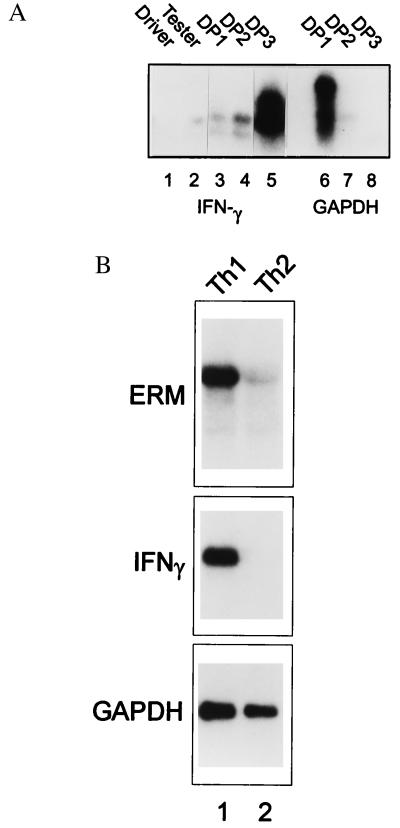
These results suggest that Stat4 may control expression of Th1-specific factors that could cooperate in regulating IFN-γ. To identify such factors, we used RDA (24) with mRNA from Th1 and Th2 cells 2 days after primary activation (Fig. 2). To verify that successive rounds of RDA subtraction amplified Th1-specific genes, we probed the representations and successive subtraction products with an IFN-γ cDNA (Fig. 2A). IFN-γ signal was detected in the Tester (Th1), but not Driver (Th2), representation and increased through subsequent subtractions DP1 (first difference product), DP2, and DP3. As a negative control, we showed that the GAPDH signal was extinguished, being virtually undetectable after the second subtraction (Fig. 2A). DP3s from DpnII and Tsp509I representations were cloned into BamHI and EcoRI pBSSKII vectors, and individual colonies were screened by dot-blot probed with Tester and Driver representation (data not shown). Colonies with differential hybridization were sequenced, and differential expression was verified by Northern blot analysis of Th1 and Th2 RNA (data not shown).
Figure 2.
Cloning of Th1-specific genes by RDA. (A) Two hundred-microgram PCR products of Driver (Th1) and Tester (Th2), as well as each difference subtraction product DP1, DP2, and DP3, were electrophoresed on 1.5% agarose gel and transferred to Zeta membrane. The membrane was probed by either IFN-γ or GAPDH cDNA probe as indicated. (B) Splenocytes from DO11.10 TCR-transgenic mice were activated with OVA and APCs with either IL-12 or IL-4, as indicated, to initiate Th1 development (lane 1) or Th2 development (lane 2) for 48 hr. Total RNA was prepared and Northern blot analysis was performed by using a full-length ERM cDNA as probe (ERM). The blot was stripped and reprobed for IFN-γ and GAPDH as indicated.
Several identified RNA were known to be selectively expressed by Th1 cells, including IFN-γ and granzyme B. However, 41 of 200 independent isolates were identical, designated D15, and were homologous to the 3′ untranslated region of the human ERM gene, a member of the PEA3 subgroup of Ets transcription factors (26). We isolated a full-length, 4-kb murine ERM cDNA from a Th1-specific cDNA library encoding a 510-aa ORF. Our clone contained three differences in amino acid sequence compared with the previously reported murine ERM cDNA, specifically 48Q>H, 271F>Y, and 283V>A (27). These differences lie outside of the ETS and transactivation domains, and 271Y and 283A are identical to the corresponding human residues (27). Murine and human ERM are highly homologous, share 95% identity at the amino acid level, and are identical within the ETS domain.
ERM mRNA was expressed at high levels in Th1 T cells and markedly reduced in Th2 cells (Fig. 2B Top), similar to the Th1 specificity shown by IFN-γ (Fig. 2B Middle). In contrast, GAPDH was expressed similarly by Th1 and Th2 cells (Fig. 2B Bottom). Consistent with the reported expression of human ERM (26), we found murine ERM expressed in brain, lung, and in lymphoid tissues, but not liver (data not shown). To confirm that our ERM cDNA was full-length and functional, we carried out in vitro translation (not shown), obtaining a single product of approximately 70 kDa, consistent with the size reported for in vitro translation of human ERM.
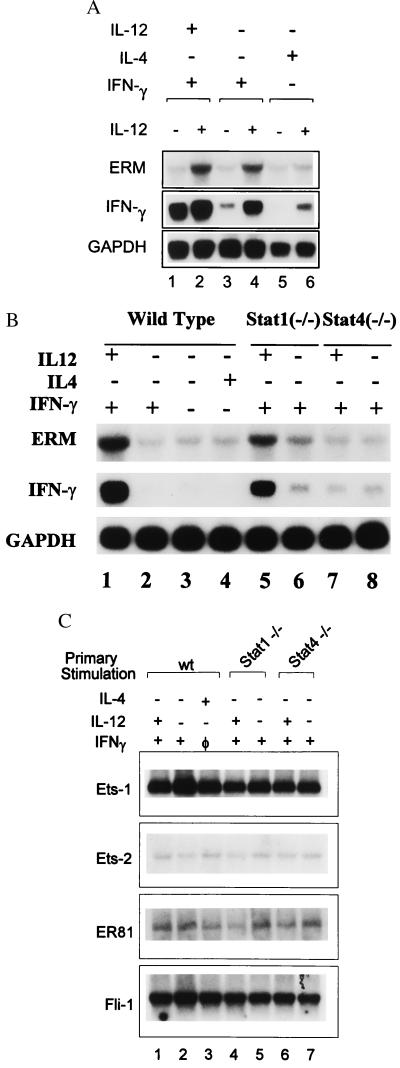
To determine the basis for Th1-specific expression of ERM, DO11.10 T cells (21) were activated with 0.3 μM of OVA under several conditions, allowed to develop for 7 days, and restimulated with and without IL-12, and ERM and IFN-γ expression was analyzed by Northern blotting after 2 days (Fig. 3A). In primary conditions known to induce IL-12 receptors (lanes 1–4) (18), IL-12 addition in secondary stimulation induced ERM. In contrast, in conditions that extinguish IL-12 receptor expression (lanes 5, 6), IL-12 addition in secondary stimulation did not induce ERM. In contrast to ERM, IFN-γ expression was IL-12-independent if Th1 cells were exposed to IL-12 in the primary culture. However, when primary cultures contained only IFN-γ and not IL-12, both ERM and IFN-γ induction required IL-12 in secondary stimulation. The results confirm that ERM is an IL-12-inducible gene.
Figure 3.
ERM expression is IL-12-inducible and Stat4-dependent. (A) Wild-type DO11.10 TCR-transgenic splenocytes were activated with OVA and APCs plus various combinations of IL-12, IL-4, and IFN-γ as indicated. On day 7, T cells were harvested and restimulated with OVA and APCs, with (+) or without (−) the addition of IL-12 (10 units/ml). Total RNA was isolated after 48 hr, and Northern blot analysis for ERM, IFN-γ, and GAPDH was performed as above. (B) DO11.10 splenocytes from wild-type, Stat1-deficient, or Stat4-deficient mice DO11.10 mice were activated by OVA in the presence (+) or absence (−) of indicated cytokines for 48 hr. RNA was prepared and Northern blot analysis of ERM, IFN-γ, and GAPDH was performed as described in A. (C) The indicated Ets family members were obtained as expressed sequence tag clones from Genome Systems (St. Louis), and their identity was confirmed by DNA sequencing. Probes were derived from unique coding or 3′ untranslated regions.
Because IL-12 activates both Stat1 and Stat4, we asked whether these factors were required for ERM induction. Stat1-deficient, Stat4-deficient, and wild-type DO11.10 TCR-transgenic T cells were activated under various primary conditions and ERM expression was analyzed by Northern blotting (Fig. 3B). ERM expression in wild-type DO11.10 T cells was dependent on addition of IL-12, but not IFN-γ or IL4 (Fig. 3B, lane 1–4). In Stat1-deficient T cells, IL-12, but not IFN-γ, induced ERM expression (Fig. 3B, lanes 5 and 6). In contrast, in Stat4-deficient T cells, IL-12 failed to induce ERM expression (lanes 7 and 8). These results suggest that ERM induction by IL-12 is dependent on Stat4, but not Stat1 (Fig. 3B). No changes in expression were seen among the cytokine conditions analyzed above for Ets-1, Ets-2, ER81, Fli-1 (Fig. 3C), or for other Ets members including PEA3, Elf-1, and PU.1 (data not shown). Thus, ERM represents the first Th1-specific transcription factor selectively induced by IL-12 through Stat4, and this pattern seems unique among the Ets transcription family.
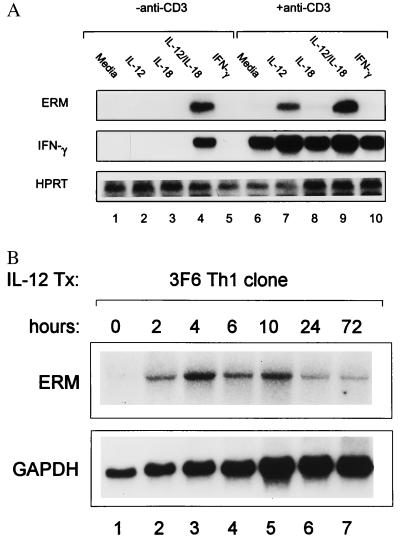
TCR signaling was considered the primary pathway for IFN-γ induction in Th1 cells, but, recently, the combination of IL-12 and IL-18 was shown to induce IFN-γ in a TCR-independent manner (28). Thus, we asked whether IL-12/IL-18 treatment also induced ERM expression (Fig. 4). Resting DO11.10 Th1 cells were activated with combinations of anti-CD3, IL-12 and IL-18, and ERM and IFN-γ expression analyzed by Northern blotting (Fig. 4). In the absence of anti-CD3 stimulation, ERM expression was induced only by the combination of IL-12 and IL-18 treatment, but not with either cytokine alone, similar to the pattern of IFN-γ expression (Fig. 4A, lanes 1–5). With anti-CD3 stimulation, ERM was induced by the addition of IL-12 (Fig. 4A, lanes 7 and 9) but not IL-18 (lane 8). In contrast, IFN-γ was induced by anti-CD3 treatment alone, without the addition of IL-12 or IL-18, and was only moderately augmented by cytokine addition. ERM is induced rapidly by IL-12 and TCR stimulation (Fig. 4B), seen after 2 hr, with the maximum between 4 and 10 hr, and is diminished significantly by 3 days. Thus, like IFN-γ, ERM can be induced by IL-12/IL-18 treatment independently of TCR signaling, but, distinct from IFN-γ, ERM cannot be induced by TCR signaling alone.
Figure 4.
Regulation of ERM expression by IL-12 and IL-18. (A) Th1 cells were generated from DO11.10 TCR-transgenic splenocytes as described above, restimulated with OVA and APCs, and, 7 days later, restimulated with the indicated combinations of anti-CD3, IL-12 (10 units/ml), IL-18 (50 ng/ml), and IFN-γ (100 units/ml) for 48 hr. Total RNA was prepared and Northern blot analysis of ERM, IFN-γ, and GAPDH was performed. (B) Resting 3F6 T cells were stimulated by using anti-CD3-coated plates and IL-12 (10 units/ml) for the indicated times, total RNA was prepared, and ERM and GAPDH Northern blot analysis was performed as described above.
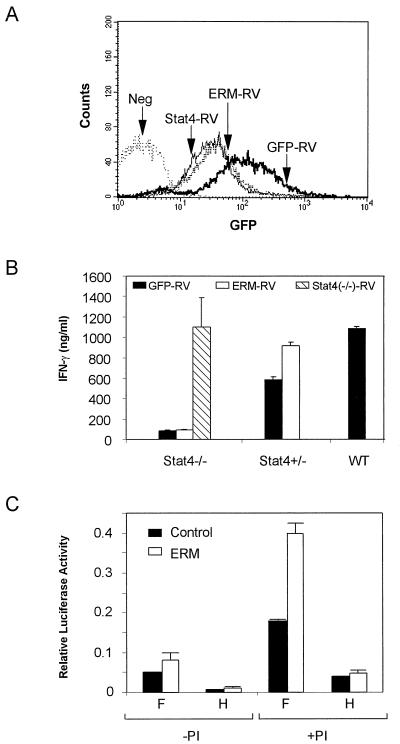
To test the role of ERM in Th1 development, we introduced ERM into Stat4-deficient, Stat4-heterozygous, and wild-type naïve DO11.10 CD4+ T cells during primary activation by using a retroviral expression system (15). Viral infection was achieved in 20–40% of primary CD4+ T cells, and infected T cells were purified by sorting for expression of GFP and CD4, expanded in vitro with OVA/APCs and IL-12 as pure, transfected populations for 7 days (Fig. 5A). T cells were restimulated with OVA and APCs, and IFN-γ production was analyzed by ELISA from 48-hr supernatants (Fig. 5B). Stat4-deficient T cells transfected with empty vector produced very low IFN-γ (Fig. 5B), as expected (8, 9). In contrast, Stat4-deficient T cells infected with Stat4-expressing retrovirus produced significantly higher IFN-γ, at levels similar to that produced by a wild-type Th1 control (Fig. 5B), confirming this system’s ability to restore a normal phenotype. However, ERM expression by retrovirus into Stat4-deficient cells did not restore normal IFN-γ production. Thus, ERM either is unrelated to IFN-γ regulation or requires cooperation with other transcription factors such as Stat4. To address this possibility, we examined Stat4-heterozygous DO11.10 T cells. First, we found that Stat4-heterozygous DO11.10 T cells exhibit a minor defect in IFN-γ production relative to wild-type controls (Fig. 5B), suggesting that Stat4 may be limiting during Th1 development. In Stat4-heterozygous T cells, retroviral expression of ERM augmented IFN-γ production to levels similar to wild-type Th1 controls (Fig. 5B). This result may suggest an interaction between ERM and Stat4, but this approach cannot distinguish between other possibilities that involve independent actions of ERM and Stat4.
Figure 5.
Effects of ERM expression on IFN-γ production in Stat4-deficient and Stat4-herterozygous T cells. (A) Stat4-deficient, Stat4-heterozygous, and wild-type DO11.10 TCR-transgenic T cells were activated in vitro by using OVA/APCs and infected on day 1 after primary activation with either empty vector (GFP-RV), Stat4-expresing retrovirus (Stat4-RV), or ERM-expressing retrovirus (ERM-RV). Infected cells were purified by cell sorting on day 7 for GFP (FL1) and CD4 expression by using anti-mouse-CD4-Phycoerythrin (PharMingen). Sorted GFP+/CD4+ T cells were reactivated with OVA, APCs, and IL-12 (see Methods), and stable transfection was confirmed by GFP expression by FACS analysis 3 days later. Data shown are single color histograms of the indicated sorted transfectant populations. The negative control (Neg) is an uninfected, Stat4-deficient DO11.10 T cell population activated concurrently under the same conditions. (B) Retrovirally infected T cells from the indicated populations described in A were harvested, washed, and restimulated at 1.25 × 105/ml with 0.3 μM of OVA and irradiated BALB/c splenocytes as APCs. Supernatants were harvested after 48 hr, and IFN-γ production was determined by ELISA. Similar results were obtained in three similar independent experiments. (C) Luciferase reporter assays were performed as described in Methods by using the IFN-γ promoter constructs F and H. Transfected cells were treated with phorbol 12-myristate 13-acetate and ionomycin (PI+) or left untreated (PI−) as described (25). Data are presented as relative light units after normalization for transfection efficiency by using CMV-Renilla luciferase.
To test for potential interaction of ERM at the IFN-γ promoter, we cotransfected ERM or control expression plasmid with IFN-γ promoter luciferase reporter constructs F and H (Fig. 5C) (16). Construct F, extending from −202 to +37, contains a potential Ets consensus between the −158 and −168 site, whereas construct H, extending from −58 to +37, lacks this site and contains only the proximal region proposed to bind AP1/ATF2 and CREB/ATF1 (29). ERM coexpression led to an approximate doubling of luciferase activity from IFN-γ promoter construct F, but had no effect on H (Fig. 5C). Thus, although ERM can influence the IFN-γ promoter in a minor way, it is unlikely to be the sole Th1-specific factor leading to IFN-γ production.
DISCUSSION
Intense efforts have been directed at understanding the molecular basis of Th1 and Th2 development (1, 2). The role of IL-12 and IL-4 in Th1 and Th2 development was recognized first in in vitro developmental systems (3, 30, 31). These systems emphasized the importance of cytokine-signaling pathways in Th1 and Th2 development, but did not reveal the underlying molecular mechanisms. Subsequent studies demonstrated that the specific transcription factors Stat4 and Stat6 mediate the phenotype-inducing effects of IL-12 and IL-4 (8–11, 32). However, it is not clear whether Stat4 and Stat6 directly mediate transcription of Th1- and Th2-specific cytokine genes or whether they act only to initiate these specific developmental programs.
Studies of IL-4 gene regulation provide some understanding of Th2 development. First, the transcription factor c-Maf was found to be Th2-specific and to interact with the proximal IL-4 promoter augmenting expression (13). Second, the transcription factor GATA-3 was shown to be Th2-specific and to augment IL-4 as well as IL-5 gene expression (14). Subsequently, the basis for Th2-selective expression of GATA-3 in T cells was shown to involve Stat6 for induction in Th2 cells and Stat4 for repression in Th1 cells (16). Whereas c-Maf acts at the MARE site in the proximal IL-4 promoter (13), GATA-3 may act at sites distant from the proximal IL-4 promoter (15), but the nature of this mechanism is not yet clear.
In contrast, less is known about the mechanisms of Th1 development. IFN-γ gene transcription appears to be under complex control and induced by at least two distinct signaling pathways (28). In Th1 cells, TCR signaling fully activates IFN-γ transpiration, but, in addition, a TCR-independent pathway exists that can be activated by the combination of cytokines IL-12 and IL-18 (28). Although many studies have aimed to identify cis-regulatory elements important for IFN-γ gene regulation, most have used transformed tumor lines and polyclonal or chemical activation, thus obscuring the role of various signaling pathways for interactions with cis-acting elements. Several transcription factors have been implicated in IFN-γ-gene regulation, including NF-AT, NFκB, YY-1, CREB/ATF1, AP-1/ATF2, GATA-3, and Stat4 (19, 33–36). Of these, none appears to be expressed in a strictly Th1-dependent manner. GATA-3, suggested previously to augment the IFN-γ promoter (36), is expressed selectively by Th2 cells, which do not produce IFN-γ, perhaps suggesting a repressive role in the IFN-γ promoter. However, we showed recently that GATA-3 does not directly inhibit IFN-γ when expressed in fully developed Th1 cells, arguing against a direct promoter effect (16). Stat4 has also been suggested to potentially regulate the IFN-γ gene (19). Several low-affinity, nonconsensus STAT sites were identified in the first intron of the IFN-γ gene, and these sites were shown to bind purified STAT proteins in vitro (19). However, sites interacting with recombinant Stat1, Stat4, as well as Stat6 were identified, and the particular role of each STAT in regulation was not established. Moreover, the functional role of these potential cis-acting elements in regulating the native IFN-γ gene has not yet been analyzed.
In contrast to direct Stat4 regulation of IFN-γ, we show here that Stat4 activation alone is not sufficient to induce IFN-γ gene transcription (Fig. 1). Because Th1 development in CD4+ T cells is dependent on Stat4 activation (8, 9), this suggested to us that Stat4 may be required to induce additional factors that may directly influence cytokine transcription. Here we report the identification of a transcription factor induced in an IL-12- and Stat4-dependent manner. This factor, ERM, is selectively expressed by Th1 cells after activation in the presence of IL-12.
ERM is a member of the Ets family of winged helix–turn–helix transcription factors, which bind the purine-rich DNA core consensus GGAA/T through a highly conserved DNA-binding domain (37). The Ets family is involved in numerous developmental processes (37, 38). The prototypical Ets factor Ets-1 selectively participates in natural killer cell development (39), and PU.1, a related Ets factor, participates in lymphoid–myeloid differentiation (40, 41). Ets transcription factors bind DNA as monomers and rely heavily on interactions with other transcription factors for exerting their unique regulatory activities (37). Importantly, human ERM binds a consensus nucleotide core sequence GGAA and can cooperate with the AP-1 family member c-Jun for transcriptional activation (42–44). ERM only recently was identified and is a member of the PEA3 subfamily of Ets proteins (44, 45). In all PEA3 family members, domains inhibiting DNA binding have been identified adjacent to the ETS domain, thought to imply that DNA binding will be regulated. However, a specific role of ERM during development has not yet been established.
This study reports a Th1-specific transcription factor specifically induced by IL-12 and Stat4. We show that expression of ERM in a Stat4-deficient T cell context is not sufficient for restoring IFN-γ production to normal Th1 levels. Thus, ERM is not likely to be the sole IL-12-induced transcription factor responsible for high IFN-γ production in Th1 cells. However, ERM overexpression augmented IFN-γ production in Stat4-heterozygous T cells. That Stat4-heterozygous T cells showed diminished IFN-γ production suggests that Stat4 may be limiting during Th1 development. Thus, ERM could cooperate with Stat4, or some Stat4-induced factor, in the regulation of IFN-γ gene transcription. ERM only minimally augments IFN-γ promoter activity in coexpression studies. However, the lack of greater augmentation implies either that relevant targets lie outside of this promoter region or that ERM requires additional factors for strong IFN-γ promoter transactivation. Thus, genetic disruption of ERM may be required to determine fully its role in Th1 development.
Acknowledgments
We thank Parveen Chand for excellent technical help and Memhet L. Guler for advice and discussions. This work was supported by National Institutes of Health Grants AIDK39676, HL56419, and AI 34580. K.M.M. is an associate of the Howard Hughes Medical Institute.
ABBREVIATIONS
- IL
interleukin
- Th
T helper
- IFN
interferon
- TCR
T cell antigen receptor
- OVA
ovalbumin peptide
- GFP
green fluorescent protein
- RDA
representational difference analysis
References
- 1.Abbas A K, Murphy K M, Sher A. Nature (London) 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 2.Seder R A, Paul W E. Annu Rev Immunol. 1994;12:635–673. doi: 10.1146/annurev.iy.12.040194.003223. [DOI] [PubMed] [Google Scholar]
- 3.Hsieh C S, Macatonia S E, O’Garra A, Murphy K M. Int Immunol. 1993;5:371–382. doi: 10.1093/intimm/5.4.371. [DOI] [PubMed] [Google Scholar]
- 4.Macatonia S E, Hosken N A, Litton M, Vieira P, Hsieh C S, Culpepper J A, Wysocka M, Trinchieri G, Murphy K M, O’Garra A. J Immunol. 1995;154:5071–5079. [PubMed] [Google Scholar]
- 5.Manetti R, Parronchi P, Giudizi M G, Piccinni M-P, Maggi E, Trinchieri G, Romagnani S. J Exp Med. 1993;177:1199–1204. doi: 10.1084/jem.177.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacobson N G, Szabo S J, Weber-Nordt R M, Zhong Z, Schreiber R D, Darnell J E, Jr, Murphy K M. J Exp Med. 1995;181:1755–1762. doi: 10.1084/jem.181.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bacon C M, Petricoin E F, III, Ortaldo J R, Rees R C, Larner A C, Johnston J A, O’Shea J J. Proc Natl Acad Sci USA. 1995;92:7307–7311. doi: 10.1073/pnas.92.16.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan M H, Sun Y L, Hoey T, Grusby M J. Nature (London) 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 9.Thierfelder W E, van Deursen J M, Yamamoto K, Tripp R A, Sarawar S R, Carson R T, Sangster M Y, Vignali D A, Doherty P C, Grosveld G C, Ihle J N. Nature (London) 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 10.Kaplan M H, Schindler U, Smiley S T, Grusby M J. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 11.Shimoda K, van Deursen J, Sangster M Y, Sarawar S R, Carson R T, Tripp RA, Chu C, Quelle F W, Nosaka T, Vignali D A, et al. Nature (London) 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 12.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Nature (London) 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 13.Ho I C, Hodge M R, Rooney J W, Glimcher L H. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 14.Zheng W, Flavell R A. Cell. 1997;89:587–596. [Google Scholar]
- 15.Ranganath S, Ouyang W J, Bhattarcharya D, Sha W C, Grupe A, Peltz G, Murphy K M. J Immunol. 1998;161:3822–3826. [PubMed] [Google Scholar]
- 16.Ouyang, W. J., Ranganath, S., Weindel, K., Bhattacharya, D., Murphy, T. L., Sha, W. C. & Murphy, K. M. Immunity9, 745–755. [DOI] [PubMed]
- 17.Szabo S J, Jacobson N G, Dighe A S, Gubler U, Murphy K M. Immunity. 1995;2:665–675. doi: 10.1016/1074-7613(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 18.Szabo S J, Dighe A S, Gubler U, Murphy K M. J Exp Med. 1997;185:817–824. doi: 10.1084/jem.185.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X, Sun Y L, Hoey T. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 20.Meraz M A, White J M, Sheehan K C, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, et al. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh C S, Macatonia S E, Tripp C S, Wolf S F, O’Garra A, Murphy K M. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh C S, Heimberger A B, Gold J S, O’Garra A, Murphy K M. Proc Natl Acad Sci USA. 1992;89:6065–6069. doi: 10.1073/pnas.89.13.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guler M L, Jacobson N G, Gubler U, Murphy K M. J Immunol. 1997;159:1767–1774. [PubMed] [Google Scholar]
- 24.Hubank M, Schatz D G. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szabo S J, Gold J S, Murphy T L, Murphy K M. Mol Cell Bio. 1993;13:4793–4805. doi: 10.1128/mcb.13.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monte D, Baert J L, Defossez P A, de Launoit Y, Stehelin D. Oncogene. 1994;9:1397–1406. [PubMed] [Google Scholar]
- 27.Chotteau-Lelievre A, Desbiens X, Pelczar H, Defossez P A, de Launoit Y. Oncogene. 1997;15:937–952. doi: 10.1038/sj.onc.1201261. [DOI] [PubMed] [Google Scholar]
- 28.Robinson D, Shibuya K, Mui A, Zonin F, Murphy E, Sana T, Hartley S B, Menon S, Kastelein R, Bazan F, O’Garra A. Immunity. 1997;7:571–581. doi: 10.1016/s1074-7613(00)80378-7. [DOI] [PubMed] [Google Scholar]
- 29.Aune T M, Penix L A, Rincon M R, Flavell R A. Mol Cell Biol. 1997;17:199–208. doi: 10.1128/mcb.17.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Gros G, Ben-Sasson S Z, Seder R A, Finkelman F D, Paul W E. J Exp Med. 1990;172:921–929. doi: 10.1084/jem.172.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manetti R, Gerosa F, Giudizi M G, Biagiotti R, Parronchi P, Piccinni M P, Sampognaro S, Maggi E, Romagnani S, Trinchieri G, et al. J Exp Med. 1994;179:1273–1283. doi: 10.1084/jem.179.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobson N G, Szabo S J, Guler M L, Gorham J D, Murphy K M. Res Immunol. 1995;146:446–456. doi: 10.1016/0923-2494(96)83014-8. [DOI] [PubMed] [Google Scholar]
- 33.Ye J, Cippitelli M, Dorman L, Ortaldo J R, Young H A. Mol Cell Biol. 1996;16:4744–4753. doi: 10.1128/mcb.16.9.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sica A, Dorman L, Viggiano V, Cippitelli M, Ghosh P, Rice N, Young H A. J Biol Chem. 1997;272:30412–30420. doi: 10.1074/jbc.272.48.30412. [DOI] [PubMed] [Google Scholar]
- 35.Barbulescu K, Becker C, Schlaak J F, Schmitt E, Meyer zum Buschenfelde K H, Neurath M F. J Immunol. 1998;160:3642–3647. [PubMed] [Google Scholar]
- 36.Penix L, Weaver W M, Pang Y, Young H A, Wilson C B. J Exp Med. 1993;178:1483–1496. doi: 10.1084/jem.178.5.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crepieux P, Coll J, Stehelin D. Crit Rev Oncog. 1994;5:615–638. [PubMed] [Google Scholar]
- 38.Bassuk A G, Leiden J M. Adv Immunol. 1997;64:65–104. doi: 10.1016/s0065-2776(08)60887-1. [DOI] [PubMed] [Google Scholar]
- 39.Barton K, Muthusamy N, Fischer C, Ting C N, Walunas T L, Lanier L L, Leiden J M. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 40.Olson M C, Scott E W, Hack A A, Su G H, Tenen D G, Singh H, Simon M C. Immunity. 1995;3:703–714. doi: 10.1016/1074-7613(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 41.Scott E W, Fisher R C, Olson M C, Kehrli E W, Simon M C, Singh H. Immunity. 1997;6:437–447. doi: 10.1016/s1074-7613(00)80287-3. [DOI] [PubMed] [Google Scholar]
- 42.Janknecht R, Monte D, Baert J L, de Launoit Y. Oncogene. 1996;13:1745–1754. [PubMed] [Google Scholar]
- 43.Laget M P, Defossez P A, Albagli O, Baert J L, Dewitte F, Stehelin D, de Launoit Y. Oncogene. 1996;12:1325–1336. [PubMed] [Google Scholar]
- 44.Nakae K, Nakajima K, Inazawa J, Kitaoka T, Hirano T. J Biol Chem. 1995;270:23795–23800. doi: 10.1074/jbc.270.40.23795. [DOI] [PubMed] [Google Scholar]
- 45.de Launoit Y, Baert J L, Chotteau A, Monte D, Defossez P A, Coutte L, Pelczar H, Leenders F. Biochem Mol Med. 1997;61:127–135. doi: 10.1006/bmme.1997.2605. [DOI] [PubMed] [Google Scholar]