Abstract
CyBase was originally developed as a database for backbone-cyclized proteins, providing search and display capabilities for sequence, structure and function data. Cyclic proteins are interesting because, compared to conventional proteins, they have increased stability and enhanced binding affinity and therefore can potentially be developed as protein drugs. The new CyBase release features a redesigned interface and internal architecture to improve user-interactivity, collates double the amount of data compared to the initial release, and hosts a novel suite of tools that are useful for the visualization, characterization and engineering of cyclic proteins. These tools comprise sequence/structure 2D representations, a summary of grafting and mutation studies of synthetic analogues, a study of N- to C-terminal distances in known protein structures and a structural modelling tool to predict the best linker length to cyclize a protein. These updates are useful because they have the potential to help accelerate the discovery of naturally occurring cyclic proteins and the engineering of cyclic protein drugs. The new release of CyBase is available at http://research1t.imb.uq.edu.au/cybase
INTRODUCTION
Proteins with a macrocyclic backbone consisting of a continuous cycle of peptide bonds have been discovered over recent years in bacteria, plants and animals (1). These macrocyclic proteins are different from small cyclic peptides, such as cyclosporin, in that they are gene-encoded products, with backbone cyclization occurring as a post-translational modification rather than being non-ribosomally synthesized (2). Currently, there are five major classes of naturally occurring cyclic proteins: the cyclic sex-pilin (3) and bacteriocins (4–6) from bacterial sources, the θ-defensins from primates (7), trypsin inhibitors from Asteraceae and Cucurbitaceae family plants (8–10) and the cyclotides from plants of the Violaceae and Rubiaceae (11–13). The cyclotides are by far the largest family of circular proteins, with recent screening programs suggesting that the total number of sequences may be in the thousands (14,15).
Interest in cyclic proteins has been inspired by the promising therapeutic advantages of cyclic proteins over their conventional linear counterparts (16–19). One of the major benefits of a circular backbone is improved stability (16,17) and at least one family of cyclic proteins, the cyclotides, has been shown to be highly resistant to enzymatic, thermal and chemical treatment (20,21). This increased stability means that circular proteins are promising scaffolds for drug design applications (22,23). The concept of backbone cyclization can be adopted to improve the bioavailability of linear proteins, thus increasing the therapeutic potential (24–27). Furthermore, rigidification of the often-flexible termini through cyclization can lead to favourable entropy changes and improved receptor binding affinities (8,9).
CyBase is a database of cyclic proteins that was initially developed to provide a uniform repository to handle the sequence/structure/function data for circular proteins (28). CyBase has now been completely redesigned to manage the continuing growth of circular protein data and to provide improved user-interactivity. A major feature of the new release is a new module to manage data on synthetic circular proteins, which was designed to assist in circular protein engineering. Additionally, a range of analytical and predictive tools has been designed to handle the unique challenges of circular protein characterization and engineering.
IMPROVEMENTS AND DISCUSSION
Although CyBase has been completely redesigned, several core features of the original CyBase release, including the underlying database architecture and the general search and display capabilities, have been retained in the new version. Information on sequence, structure and function is stored in a MySQL database, where the central protein table, which contains information on each characterized cyclic protein, is linked to additional tables that described nucleic acid sequences, structures, activities and literature references. The data is accessed using a web-based interface, which provides a variety of text- or alignment-based searching methods. Data entries are displayed using dynamically generated data cards, which describe the relevant information, including sequence, classification and cross-links to other entries in CyBase or to external biological databases such as Genbank, UniProt and PDB. In the original CyBase release, the interface was adapted from a popular content management system for community websites written using the PHP language. In the new release, the interface has been substantially redesigned to increase user-interactivity and improve integration of data with tools. These improvements have been achieved using an additional data abstraction layer implemented in XML that also improves the extensibility and maintainability of the database.
As of August 2007, CyBase includes 251 protein sequences, 49 nucleic acid sequences, 39 structures and 91 activity-related entries from five classes of circular proteins. The data content of CyBase is now almost double the initial release, and the growth is expected to continue, with a recent study suggesting that in at least one family of circular proteins, the cyclotides, >9000 sequences have yet to be characterized (14). In addition, an increasing number of engineering studies are being applied to circular proteins.
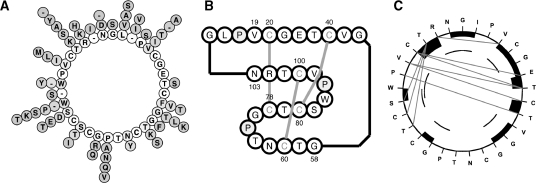
The new CyBase release provides a new range of tools to aid in cyclic protein visualization, discovery and engineering. In terms of visualization, the ‘Diversity Wheel’ tool generates a novel representation of circular protein sequence variation. The tool accepts a multiple sequence alignment and generates a wheel-like diagram that is composed of an inner circle, which describes the consensus sequence from the given multiple sequence alignment, and the radial spikes from each position represent the different amino acids observed at that position, as shown in Figure 1. This representation is useful for evolutionary or mutational studies of circular proteins. For cyclotides and squash trypsin inhibitors, a ‘Collier de Perles’ graphical representation (29) of the sequence/structure has been adapted from the KNOTTIN database (30) to handle the cyclic nature of cyclotides. An example ‘Collier de Perles’ representation is shown in Figure 1. This representation provides a link between protein sequences and their structures and is particularly useful for protein engineering, sequence–structure analysis, visualization and comparisons of positions for mutations, polymorphisms and contact analysis (29). For the visualization of structures, a tool based on Jmol (http://www.jmol.net) has been added to the structure cards to allow a quick overview of each structure and to highlight crucial structural features such as the surface hydrophobicity. In combination with the activity entries in CyBase, visualization of the structures assists in identifying structure–activity relationships.
Figure 1.
Sequence graphical representations incorporated into CyBase. Panel (A) shows a Diversity Wheel representation of sequence diversity from a multiple sequence alignment, where the consensus sequence is positioned in the inner circle and the spike protruding from each position represents the amino acid variation observed at that position. Panel (B) is a Collier de Perles representation of the prototypical cyclotide, kalata B1, showing the sequence and disulphide connectivity. Collier de Perles representations can be generated for proteins belonging to the cyclotide or trypsin squash inhibitor classes. Panel (C) shows a Cyclic Seqplot, which is a representation for NOE data measured from an NMR experiment. The sequence of the peptide is shown on the outside of the circle. Backbone NOEs are drawn as dark bars, where the height of the bar is relative to the strength of the NOE. Medium range and long NOEs are drawn as arcs and lines. 134 × 47mm (600 × 600 DPI)
Several tools have been added to CyBase to facilitate cyclic protein discovery. Characterization of cyclic proteins has benefited from approaches in molecular biology and mass spectrometry (to determine sequence information) and NMR (to determine 3D structures). To assist in molecular screening for cyclic protein genes, the CyBase ‘Primer Match’ tool, which was developed from suggestions from users, can rapidly predict primer-binding sites given a list of primer sequences and a template sequence. Identification of cyclic protein genes is important because backbone cyclization is a ‘seamless’ process, which means that the location of the N- and C-termini cannot be determined from the mature peptide alone. Mass spectrometry methods, which measure the mass of the mature peptide or enzyme-digested fragments, are commonly used for rapid protein sequence determination. Existing computational tools, which form the core of protein sequence proteomics, do not consider the effect of cyclization on a protein of interest, which changes the mass of the mature protein, the pI of the protein and introduces additional fragments when the protein is digested. Accordingly, the CyBase ‘Digest Peptide’ tool allows for the in silico enzyme digestion of cyclic peptides as well as the prediction of properties such as the absorption coefficient and the pI. The CyBase ‘Fingerprint Search’ tool gives the capability to search the entire database using masses of peptide fragments obtained from an enzymatic digestion of the reduced original peptide for rapid protein sequence identification. Analysis of NMR data, such as chemical shift and NOE patterns, can provide an early indication of the structure of a protein. Chemical shifts and NOE restraints are stored in CyBase and can be presented visually for analysis and comparison. The ‘Alphaplot’ tool can easily generate chemical shift index plots, which are commonly used to identify secondary structure (31). The CyBase ‘Cyclic Seqplot’ tool offers a new representation for short- and long-range NOE patterns, which uses a circular template as shown in Figure 1, and can be used to quickly identify structural elements (e.g. secondary structure).
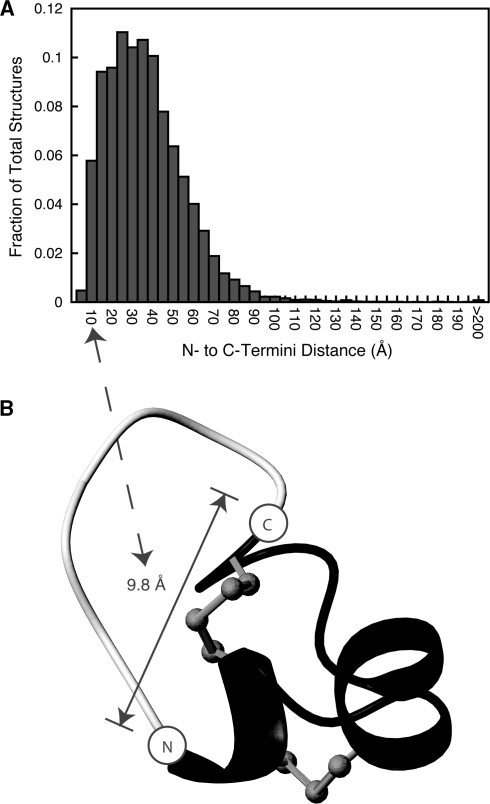
CyBase provides tools to facilitate the engineering of cyclic proteins. To help identify potential targets for backbone cyclization, the CyBase ‘Termini Distance Distributions’ page provides current statistics on N- to C-termini distances of proteins from the PDB. The distribution of distances from the PDB as of June 2007 is shown in Figure 2 and indicates that a significant number of proteins have N- to C-termini distances below 20 Å, a distance that may only require linkers made of a few residues (32). The distributions of the distances can be compared to random models. The details of two random models—one based on an ellipsoid and the other on a random-walk algorithm—have been described previously (33). The current study is further useful because the proximity of the N- and C-termini of proteins has been implicated as an important factor in protein stability and folding (34). The CyBase ‘Predict Linker’ tool predicts the size of a poly-alanine linker needed to connect the termini of a given protein. The algorithm models the cyclized structure using an increasingly longer closing linker while avoiding steric clashes by using the MODELLER program (35). An example model of an artificially cyclized protein is shown in Figure 2. Further analysis of the effect of cyclization can be made using the ‘Cyclization Energy’ tool, which predicts the change in the unfolding free energy, ΔΔGcycl, by backbone cyclization. The algorithm used for the energy prediction is based on the probability of a given linker length stretching a particular distance over the folded and unfolded states of the protein, and has been described in detail previously (36). As circular proteins have been shown to be relatively stable, circular proteins present themselves as promising scaffolds for grafting applications. By following the CyBase ‘Synthetic Analogues’ tool, users can view summaries of grafted or modified peptides and identify which variants had been successfully folded and which variants had interesting activity. The collation of this information is potentially very useful for developing rules for future studies involving synthetic cyclic peptides.
Figure 2.
Cyclization tools incorporated into CyBase. By scanning the distribution of N- to C-termini distances from the PDB as shown in panel (A), the conotoxin MII was identified as a potential target for backbone cyclization. Its relatively short N- to C-termini distance of 9.8 Å means that it is potentially more amenable to backbone cyclization compared to a protein with a longer termini distance. Panel (B) shows a model of a cyclic MII using its native linear structure as a template (PDB ID: 1MII) (37), which has been cyclized in silico using a seven-residue poly-alanine linker (coloured in white). 83 × 136 mm (600 × 600 DPI)
CONCLUSION
Cyclic proteins are interesting because they offer increased stability compared to conventional proteins and are promising drug scaffolds. CyBase is a database dedicated to cyclic proteins that provides a standardized method for accessing information on proteic sequences, nucleic sequences, 3D structures and assay results. CyBase also manages data on synthetic analogues of cyclic proteins to assist in drug development projects. Since its initial release, CyBase has grown in size and now provides a suite of tools that are useful for the visualization, analysis and characterization and engineering of cyclic proteins. These include a new ‘Diversity Wheel’ representation, which is useful for analysing circular protein sequence variation, and a ‘Predict Linker’ tool to help in the engineering of cyclic proteins from linear targets. CyBase is available at http://research1t.imb.uq.edu.au/cybase/.
ACKNOWLEDGEMENTS
The authors thank Dr Huan-Xiang Zhou for helping with the development of the ‘Cyclization Energy’ tool used to predict the change in unfolding free energy by backbone cyclization. Funding to pay the Open Access publication charges for this article was provided by Australian Research Council and National Health and Medical Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Trabi M, Craik DJ. Circular proteins: no end in sight. Trends Biochem. Sci. 2002;27:132–138. doi: 10.1016/s0968-0004(02)02057-1. [DOI] [PubMed] [Google Scholar]
- 2.Kohli RM, Walsh CT. Enzymology of acyl chain macrocyclization in natural product biosynthesis. Chem. Commun. 2003;3:297–307. doi: 10.1039/b208333g. [DOI] [PubMed] [Google Scholar]
- 3.Eisenbrandt R, Kalkum M, Lai EM, Lurz R, Kado CI, Lanka E. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 4.Kawai Y, Saito T, Toba T, Samant S, Itoh T. Isolation and characterization of a highly hydrophobic new bacteriocin (gassericin A) from Lactobacillus gasseri LA39. Biosci. Biotechnol. Biochem. 1994;58:1218–1221. doi: 10.1271/bbb.58.1218. [DOI] [PubMed] [Google Scholar]
- 5.Kemperman R, Kuipers A, Karsens H, Nauta A, Kuipers O, Kok J. Identification and characterization of two novel clostridial bacteriocins, circularin A and closticin 574. Appl. Environ. Microbiol. 2003;69:1589–1597. doi: 10.1128/AEM.69.3.1589-1597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maqueda M, Galvez A, Bueno MM, Sanchez-Barrena MJ, Gonzalez C, Albert A, Rico M, Valdivia E. Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr. Protein. Pept. Sci. 2004;5:399–416. doi: 10.2174/1389203043379567. [DOI] [PubMed] [Google Scholar]
- 7.Tang YQ, Yuan J, Osapay K, Tran D, Miller CJ, Ouellette AJ, Selsted ME. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated α-defensins. Science. 1999;286:498–502. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- 8.Luckett S, Garcia RS, Barker JJ, Konarev AV, Shewry PR, Clarke AR, Brady RL. High resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds. J. Mol. Biol. 1999;290:525–533. doi: 10.1006/jmbi.1999.2891. [DOI] [PubMed] [Google Scholar]
- 9.Korsinczky ML, Schirra HJ, Rosengren KJ, West J, Condie BA, Otvos L, Anderson MA, Craik DJ. Solution structures by 1H NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant. J. Mol. Biol. 2001;311:571–591. doi: 10.1006/jmbi.2001.4887. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez JF, Gagnon J, Chiche L, Nguyen TM, Andrieu JP, Heitz A, Hong TT, Pham TT, Nguyen DL. Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry. 2000;39:5722–5730. doi: 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
- 11.Craik DJ, Daly N, Mulvenna J, Plan M, Trabi M. Plant cyclotides: a unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 12.Jennings C, West J, Waine C, Craik D, Anderson M. Biosynthesis and insecticidal properties of plant cyclotides: the cyclic knotted proteins from Oldenlandia affinis. Proc. Natl Acad. Sci. USA. 2001;98:10614–10619. doi: 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goransson U, Svangard E, Claeson P, Bohlin L. Novel strategies for isolation and characterization of cyclotides: the discovery of bioactive macrocyclic plant polypeptides in the Violaceae. Curr. Protein Pept. Sci. 2004;5:317–329. doi: 10.2174/1389203043379495. [DOI] [PubMed] [Google Scholar]
- 14.Craik DJ, Daly NL, Mulvenna J, Plan MR, Trabi M. Discovery, structure and biological activities of the cyclotides. Curr. Protein Pept. Sci. 2004;5:297–315. doi: 10.2174/1389203043379512. [DOI] [PubMed] [Google Scholar]
- 15.Simonsen SM, Sando L, Ireland DC, Colgrave ML, Bharathi R, Goransson U, Craik DJ. A continent of plant defense peptide diversity: cyclotides in Australian Hybanthus (Violaceae) Plant Cell. 2005;17:3176–3189. doi: 10.1105/tpc.105.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwai H, Pluckthun A. Circular beta-lactamase: stability enhancement by cyclizing the backbone. FEBS Lett. 1999;459:166–172. doi: 10.1016/s0014-5793(99)01220-x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou HX. Loops, linkages, rings, catenanes, cages, and crowders: entropy-based strategies for stabilizing proteins. Acc. Chem. Res. 2004;37:123–130. doi: 10.1021/ar0302282. [DOI] [PubMed] [Google Scholar]
- 18.Felizmenio-Quimio ME, Daly NL, Craik DJ. Circular proteins in plants – solution structure of a novel macrocylic trypsin inhibitor from Momordica cochinchinensis. J. Biol. Chem. 2001;276:22875–22882. doi: 10.1074/jbc.M101666200. [DOI] [PubMed] [Google Scholar]
- 19.Katsara M, Tselios T, Deraos S, Deraos G, Matsoukas MT, Lazoura E, Matsoukas J, Apostolopoulos V. Round and round we go: cyclic peptides in disease. Curr. Med. Chem. 2006;13:2221–2232. doi: 10.2174/092986706777935113. [DOI] [PubMed] [Google Scholar]
- 20.Gran L, Sandberg F, Sletten K. Oldenlandia affinis (R&S) DC. A plant containing uteroactive peptides used in African traditional medicine. J. Ethnopharmacol. 2000;70:197–203. doi: 10.1016/s0378-8741(99)00175-0. [DOI] [PubMed] [Google Scholar]
- 21.Colgrave ML, Craik DJ. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry. 2004;43:5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 22.Craik DJ, Cemazar M, Wang CK, Daly NL. The cyclotide family of circular miniproteins: nature's combinatorial template. Biopolymers. 2006;84:250–266. doi: 10.1002/bip.20451. [DOI] [PubMed] [Google Scholar]
- 23.Craik DJ, Clark RJ, Daly NL. Potential therapeutic applications of the cyclotides and related cystine knot mini-proteins. Expert Opin. Investig. Drugs. 2007;16:595–604. doi: 10.1517/13543784.16.5.595. [DOI] [PubMed] [Google Scholar]
- 24.Deechongkit S, Kelly JW. The effect of backbone cyclization on the thermodynamics of beta-sheet unfolding: stability optimization of the PIN WW domain. J. Am. Chem. Soc. 2002;124:4980–4986. doi: 10.1021/ja0123608. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi H, Arai M, Takenawa T, Sota H, Xie QH, Iwakura M. Stabilization of hyperactive dihydrofolate reductase by cyanocysteine-mediated backbone cyclization. J. Biol. Chem. 2007;282:9420–9429. doi: 10.1074/jbc.M610983200. [DOI] [PubMed] [Google Scholar]
- 26.Clark RJ, Fischer H, Dempster L, Daly NL, Rosengren KJ, Nevin ST, Meunier FA, Adams DJ, Craik DJ. Engineering stable peptide toxins by means of backbone cyclization: stabilization of the alpha-conotoxin MII. Proc. Natl Acad. Sci. USA. 2005;39:13767–13772. doi: 10.1073/pnas.0504613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovelace ES, Armishaw CJ, Colgrave ML, Wahlstrom ME, Alewood PF, Daly NL, Craik DJ. Cyclic MrIA: a stable and potent cyclic conotoxin with a novel topological fold that targets the norepinephrine transporter. J. Med. Chem. 2006;49:6561–6568. doi: 10.1021/jm060299h. [DOI] [PubMed] [Google Scholar]
- 28.Mulvenna J, Wang C, Craik DJ. CyBase: a database of cyclic protein sequence and structure. Nucleic Acids Res. 2006;34:D192–D194. doi: 10.1093/nar/gkj005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaas Q, Lefranc MP. IMGT Colliers de Perles: standardized sequence-structure representations of the IgSF and MhcSF superfamily domains. Curr. Bioinformatics. 2007;2:21–30. [Google Scholar]
- 30.Gelly JC, Gracy J, Kaas Q, Le-Nguyen D, Heitz A, Chiche L. The KNOTTIN website and database: a new information system dedicated to the knottin scaffold. Nucleic Acids Res. 2004;32:D156–D159. doi: 10.1093/nar/gkh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wishart DS, Sykes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 32.Chiche L, Heitz A, Gelly J-C, Gracy J, Chau PTT, Ha PT, Hernandez J-F, Le-Nguyen D. Squash inhibitors: from structural motifs to macrocyclic knottins. Curr. Protein Pept. Sci. 2004;5:341–349. doi: 10.2174/1389203043379477. [DOI] [PubMed] [Google Scholar]
- 33.Thornton JM, Sibanda BL. Amino and carboxy-terminal regions in globular proteins. J. Mol. Biol. 1983;167:443–460. doi: 10.1016/s0022-2836(83)80344-1. [DOI] [PubMed] [Google Scholar]
- 34.Krishna MMG, Englander SW. The N-terminal to C-terminal motif in protein folding and function. Proc. Natl Acad. Sci. USA. 2005;102:1053–1058. doi: 10.1073/pnas.0409114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou HX. Effect of backbone cylization on protein folding stability: chain entropies of both the unfolded and the folded states are restricted. J. Mol. Biol. 2003;332:257–264. doi: 10.1016/s0022-2836(03)00886-6. [DOI] [PubMed] [Google Scholar]
- 37.Hill JM, Oomen CJ, Miranda LP, Bingham JP, Alewood PF, Craik DJ. Three-dimensional solution structure of alpha-conotoxin MII by NMR spectroscopy: effects of solution environment on helicity. Biochemistry. 1998;37:15621–15630. doi: 10.1021/bi981535w. [DOI] [PubMed] [Google Scholar]