Abstract
The NIAID-funded Biodefense Proteomics Resource Center (RC) provides storage, dissemination, visualization and analysis capabilities for the experimental data deposited by seven Proteomics Research Centers (PRCs). The data and its publication is to support researchers working to discover candidates for the next generation of vaccines, therapeutics and diagnostics against NIAID's Category A, B and C priority pathogens. The data includes transcriptional profiles, protein profiles, protein structural data and host–pathogen protein interactions, in the context of the pathogen life cycle in vivo and in vitro. The database has stored and supported host or pathogen data derived from Bacillus, Brucella, Cryptosporidium, Salmonella, SARS, Toxoplasma, Vibrio and Yersinia, human tissue libraries, and mouse macrophages. These publicly available data cover diverse data types such as mass spectrometry, yeast two-hybrid (Y2H), gene expression profiles, X-ray and NMR determined protein structures and protein expression clones. The growing database covers over 23 000 unique genes/proteins from different experiments and organisms. All of the genes/proteins are annotated and integrated across experiments using UniProt Knowledgebase (UniProtKB) accession numbers. The web-interface for the database enables searching, querying and downloading at the level of experiment, group and individual gene(s)/protein(s) via UniProtKB accession numbers or protein function keywords. The system is accessible at http://www.proteomicsresource.org/.
INTRODUCTION
Systems approaches are increasingly being used to understand gene/protein functions and complex regulatory processes on a global scale (1). Proteomics addresses identification, profiling and structure/function of proteins at a cellular or organism level (2,3). Transcriptomics is widely used for studying genome-wide gene expression patterns and regulatory networks. Storing, disseminating and integrating these heterogeneous types of data are critical to facilitate data exchange and analysis (4–7).
There are publicly available databases for storing and disseminating proteomics or transcriptomics data, such as ArrayExpress, GEO, PRIDE, PeptideAtlas, Protein Data Bank and Global Proteomics Machine database (8–13). Most of these data repositories host individual data types and do not provide organism-wide integration of genomic, transcriptomics and proteomic data, which is essential for developing a pathosystem-centric resource needed for supporting the research community.
To facilitate community research for discovery of candidates for the next generation of vaccines, therapeutics and diagnostics, the National Institute of Allergy and Infectious Diseases (NIAID) has funded research to characterize pathogen proteomes and pathogen:host interactions, and mechanisms of pathogenesis, which includes contracts to seven PRCs that generate diverse experiment data sets from multiple pathosystems, and a Biodefense Proteomics Resource Center (RC) to store the data, provide visualization and analysis tools, and make it publicly accessible (for a complete list of organisms under investigation see the RC home page http://www.proteomicsresource.org/).
Towards this goal, the RC is hosted across three institutions (SSS, VBI, PIR) and includes a variety of information and tools covering the organisms, reagents, publications, operating procedures, protein annotations, experiment data and more. These are highly linked to maximize the value to the research community. The remainder of this article will focus on one aspect of the RC, the public proteomics repository system which was developed with the following main objectives: (i) manage and disseminate transcriptomic and proteomic data; (ii) develop a cyberinfrastructure (http://www.nsf.gov/od/oci/reports/toc.jsp) for integration and interoperability of diverse data sets. The RC is a unique publicly available proteomics data resource that hosts a wide range of ‘omics’ data sets on pathogen and host interactions and integrates all experiment data submitted by PRCs to illustrate gene or protein functions involved in pathogen biology, and host and pathogen interaction.
DATABASE AND DATA DESCRIPTION
Database architecture and application
The RC database application housing experiment data uses J2EE technologies and a N-tier architecture. The application has been modeled using Unified Modeling Language (UML) methodology.
The relational database is hosted on Oracle 9i. Data is distributed over three database instances which store experiment, protein and administrative data. Navigation between the experiment and protein databases is enabled by the use of UniProt accession numbers. Within the experiment data instance, query performance is optimized by using materialized views, which pre-join complex queries and reduce query response times.
The experiment data model includes five topic areas: (i) researcher information; (ii) protocols; (iii) experiment design and technologies; (iv) experiment results; and (v) annotation data. The database model supports multiple data types from transcriptomics, proteomics and genomics experiments. Common features across experiment types, such as experiment metadata and sample attributes, are modeled in generic data structures while experiment specific details, such as mass spectrometry charge and protein interactions, are tracked in specialized data structures. The database schema is available at the web link: http://proteinbank.vbi.vt.edu/ProteinBank/RC_database_schema.pdf.
At the middle tier, data objects and business logic are implemented using the Struts framework, a Model-View-Controller design pattern. An advantage of this approach is that it provides application developers with an abstract representation of the underlying data model which minimizes dependencies between the data model and application code.
At the front end, dynamic web pages are created by using Java Server Pages and Java Servlets.
Data integration
Data is integrated in a protein-centric manner by mapping all proteins and genes in the experimental results to UniProtKB (14) or UniParc (15) accession numbers using the id-mapping mechanism provided by the iProClass (16) system. In rare cases, RC created identifiers for gene(s)/protein(s) that could not be mapped to the existing databases. The original IDs used by the research centers are preserved. In this way every gene/protein is assigned a unique accession number which links the experimental results from the biodefense research centers to functional annotation and information from 90 biological databases, including databases for protein families, functions and pathways, interactions, structures and structural classifications, genes and genome data, ontologies, literature and taxonomy. Data integration enhances the search functionality of the system, as protein attributes from all these other sources are made available in addition to those provided by the research centers, allowing complex searches across multiple experiments and data types. Hyperlinks to external data resources are provided.
Available data
The currently available data sets and data types, reagents and the corresponding organisms at the RC are listed in Table 1.
Table 1.
Currently avaliable data sets and data types and the corresponding organisms at the RC
| Proteomics research center | Pathosystem | Experiment design and technology | Datasets/data type | Reagent |
|---|---|---|---|---|
| Caprion Proteomics Inc. | Brucella abortus | To measure the impact of BvrR/BvrS on cell envelope proteins, Caprion Proteomics Inc. has performed a label-free mass spectrometry-based proteomic analysis of spontaneously released outer membrane fragments from four strains of B. abortus. Currently, 167 outer membrane proteins were identified as interesting targets and released on the RC website. | 1 (mass spectrometry) | |
| Einstein Biodefense Proteomic Research Center | Toxoplasma gondii Cryptosporidium parvum | Apicomplexan cytoskeletal assemblies and outer membrane proteins from T. gondii and C. parvum were isolated and determined through proteomics-based methods. Currently, about 700 proteins from C. parvum and 2400 proteins from T. gondii have been identified and released on the RC website. | 2 (mass spectrometry) | Antibodies |
| Harvard Institute of Proteomics | Bacillus anthracis Vibrio cholerae | Full-length open reading frame (ORF) clones representing the complete proteome for V. cholerae and B. anthracis in protein expression-ready format are made available. These clones can be searched, ordered through the website and directly used for making protein microarrays representing the proteomes for V. cholerae and B. anthracis (32). | 3 (genomic cloning) | Clone reagent |
| Myriad Genetics, Inc. | Bacillus anthracis Yersinia pestis Homo sapiens | Protein–protein interaction maps between the human proteome and the proteomes of Category-A pathogens, B. antharcis and Y. pestis and F. tularensis, were carried out through random two-hybrid screening and directed screening technologies. Two data sets using directed screened interactions among 67 proteins from Homo sapiens and 2 proteins from B. anthracis and 4 proteins of Y. pestis were released on the website. | 2 (yeast two-hybrid system) | Clone reagent |
| Pacific Northwest National Laboratory | Salmonella typhimurium, Mus muculus | Protein abundance profile of S. typhimurium has been extensively studied using proteomics technologies in vitro using cultures grown under different life cycles, e.g. log, magnesium depletion phase and in vivo, mouse macrophages infection conditions (33–35). The data is published on the website. | 3 (mass spectrometry) | Bacteria |
| Scripps Research Institute | SARS-CoV | Is attempting to deliver a functional and structure catalog of the SARS-CoV proteome in order to initiate a comprehensive program for therapeutic intervention. Several proteins and protein domains of SARS have been determined by using NMR and/or X-ray crystallography technologies (36–41). | 11 (NMR and/or X-ray) | Clone reagent |
| University of Michigan | Bacillus anthracis Mus muculus | Protein and gene expression profile of B. anthracis have been extensively studied in vitro using cultures grown under different life cycles, e.g. different time points, and in vivo, mouse macrophages infection conditions (42–44). | 4 (microarray and mass spectrometry) | Array chip |
Besides the published data described earlier, experimental data sets, including technologies and protocols that are adopted for generating those data, continue to be submitted to the center and are being processed for public dissemination. The predicted complete proteomes of organisms, as well as the annotation data extracted from the iProClass database, are available at the link (http://www.proteomicsresource.org/Resources/Catalog.aspx).
DATA DISSEMINATION
All data stored in the RC are publicly available for query through the web navigation system at http://www.proteomicsresource.org/ or for downloading from the FTP site at ftp://141.161.76.88/pub/proteomics_ftp/. Currently, available data is summarized in the Project Catalog page (http://www.proteomicsresource.org/Resources/Catalog.aspx). From the catalog table a user can navigate to the experiment data (http://proteinbank.vbi.vt.edu/ProteinBank/g/data.dll), related publications or experimental protocols. Users can also search the integrated data and annotations in a protein centric manner (http://pir.georgetown.edu/cgi-bin/textsearch_cat.pl?search=1).
Data export
The RC supports data export at different levels, for instance: (i) summary data at organism level can be exported in different formats (e.g. FASTA), by selecting the relevant organism in the organism field of the annotation pages (http://pir.georgetown.edu/cgi-bin/textsearch_cat.pl). (ii) Data from individual experiments (e.g. identified protein list of Salmonella typhimurium grown under log phase) can be queried from the experiment data pages of mass spectrometry data type, with the experiment ID ‘PNNL_MS_SAM_05’ (http://proteinbank.vbi.vt.edu/ProteinBank/g/findexpbyid.do?id=PNNL_MS_SAM_05) and exported as a tab delimited file. (iii) Specific individual or group gene(s)/protein(s) in which the user is interested can be searched by entering keyword(s) or UniProtKB ID(s), and the search results can be exported as well. (iv) Experimental results data provided by the PRCs can be downloaded from the FTP site.
DATA SEARCH, ANALYSIS AND VISUALIZATION TOOLS
The RC not only stores, integrates and disseminates data, but also provides data visualization and analysis tools. The RC allows Boolean searches of all proteins and experimental results and provides options for batch retrieval of data by a large variety of protein-related identifiers (http://pir.georgetown.edu/pirwww/proteomics/index.shtml#MPD). In addition, a variety of protein analysis tools are provided to allow further analysis of search results (e.g. BLAST, peptide match, etc.). Search results are linked to the underlying experiment data allowing data type specific analysis and visualization. To illustrate these capabilities, two data analysis tools are described subsequently.
Protein 3D structure visualization
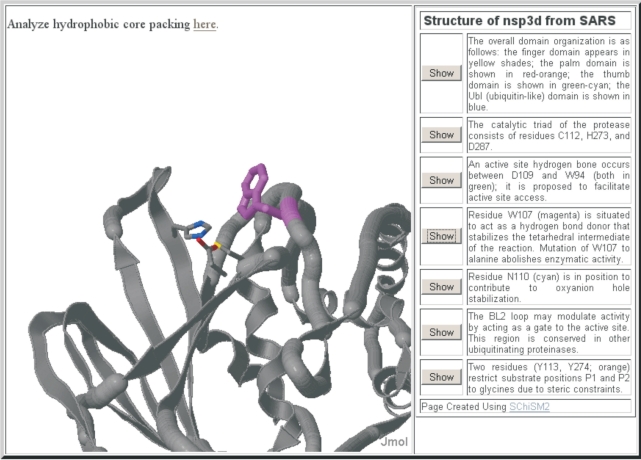
The RC provides a web-based protein-structure visualization and analysis tool (Figure 1). The tool allows visualizing the protein structure and provides the researcher with annotations derived from the features described in the publication for the protein. Multiple scenes have been illustrated for each SARS protein structure using a web-based tool that assists in designing and generating web page annotations (17). The annotations also link to a tool for interactive analysis of a protein structure or protein complexes in real-time 3D. A researcher may analyze SARS protein structures or choose to analyze any of those available from the Protein Data Bank, as well as structure files uploaded through the browser.
Figure 1.
Visualization of 3D structure of SARS-CoV PLP protease (nsp3d). The key active site residues of PLP, and a nearby tryptophan proposed to stabilize the tetrahedral intermediate in the catalytic cycle, are illustrated in the annotation for the 3D structure that is viewable at RC. The 3D structure is fully interactive and different views are obtained by clicking on the buttons associated with the views’ description. The different views illustrate features described by Ratia et al. (31).
GO term analysis
In order to support gene ontology (GO) term analysis, the publicly available AmiGO tool has been integrated with the RC system. AmiGO provides an interface to search and browse the ontology and annotation data provided by the GO consortium (http://www.geneontology.org/GO.tools.shtml). A database of GO terms, for organisms listed in Table 1, has been built into the RC system. Experimental data is seamlessly passed to the AmiGO search engine from which a GO hierarchy diagram is generated, and a GO term result frequency diagram, developed by the RC, is returned that provides the user with an overview of the GO terms. For example, the gene group from the experiment ID ‘UOM_MA_07’, as mentioned in the Data export section earlier, can be submitted for AmiGO analysis using the ‘GO analysis’ button at the bottom of the page. The frequency diagram is hyperlinked in the table header.
PROTEOMICS DATA RESOURCE APPLICATION
The RC provides the scientific community with integrated, heterogeneous, experimental data and comprehensive protein annotation, addressing pathogen life cycle biology, host response and the interaction between host and pathogen. To obtain specific experimental data, a user can navigate the RC website following the web links. For querying specific gene/protein information, the user can query the database by using the ‘site search’ function located at the top header bar of every page or the specifically designed search functionality found in the annotation and experiment data pages. In the following text, two use cases illustrate how the RC resource can be used by the scientific community.
Use case 1: search for a mouse gene responding to pathogen infection
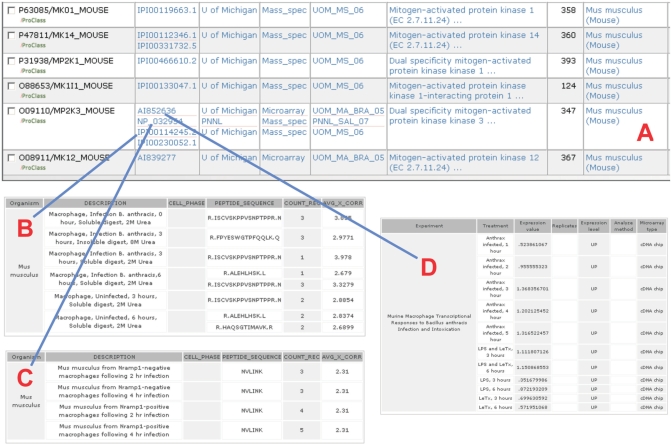
In the search page, http://pir.georgetown.edu/cgi-bin/textsearch_cat.pl, the user can query the summarized gene/protein information across multiple experiments by entering any recognized gene/protein identifier (e.g. GenBank/EMBL/DDBJ, UniProtKB accession numbers), protein names, gene names or functional keyword(s). Searching over 40 fields across the tables in the database is supported. For example, by entering the text ‘mitogen-activated’, selecting ‘protein name’ in the category field and submitting the search, a summary table of mouse ‘mitogen-activated’ protein information is presented (Figure 2A). The table can be customized with ‘Display Options’. In the page of summarized mitogen-activated proteins, it is shown that ‘mitogen-activated protein’ was detected in the mass spectrometry experiment when the macrophage was infected by Bacillus anthracis (Figure 2B) or S. typhimurium (Figure 2C). Gene expression patterns of macrophage grown with different treatments were addressed as well (Figure 2D). By following the hyperlink on the iProClass image located at the left side of Figure 2A, the user can navigate to the comprehensive annotation data of the mitogen-activated protein, such as KEGG pathway description, KEGG ID, literature and so on.
Figure 2.
Experiment and annotation data of Mouse Mitogen-activated gene. (A) search result; (B) mitogen-activated protein profile of macrophages under B. anthracis infection; (C) mitogen-activated protein profile of Nramp1-posititve and Nramp1-negative macrophages under S. typhimurium infection; (D) mitogen-activated gene expression profile of macrophages under different treatments.
Use case 2: search for organism-centric experiment data
From the Organisms page (http://proteinbank.vbi.vt.edu/ProteinBank/g/data.dll), selecting ‘Organism’ from the left navigation panel allows the user to query summarized experiment data that correspond to a specific pathosystem. For instance, all experiments carried out with B. anthracis are listed by selecting that pathosystem and submitting the query. The resulting page shows an overview of each individual experiment and allows the user to navigate all the way down to individual gene/protein information. The user can also start at the individual protein level and navigate to the experiments containing data for them. Starting at http://pir.georgetown.edu/cgi-bin/textsearch_cat.pl and using the ‘Select an Organism to Show’ drop down menu to choose Bacillus anthracis, all genes/proteins from the organism data will be listed with rich annotation. From there summarized data can be exported, tools such as BLAST can be run on individual or sets of proteins, the user can navigate to ‘experiment summaries’ by clicking on Experiment ID to find any experiments containing data on that protein, or the user can go directly to the experiment data on that individual protein by clicking on Dir.ID.
DISCUSSION
The goal of the Biodefense Proteomics Program funded by the NIAID is to generate and make publicly available the experimental data from characterization of the pathogen proteome, pathogen and host interactions, mechanisms of microbial pathogenesis, and selected host innate and adaptive immune responses to infectious agents. It is anticipated that this proteomics program will provide a research resource to the scientific community to discover potential candidates for the next generation of vaccines, therapeutics and diagnostics. Integrated and annotated experiment data in the RC provides the capability for researchers to query, visualize, download or further analyze the data to systematically study pathogenesis and host response across diverse data types and organisms.
Researchers have realized the importance of integrating proteomics, transcriptomics, genetics and metabolite data to interpret and predict gene function, complex regulatory mechanisms and to discover targets and biomarkers (4,6,18,19). In addition, open source software systems have been developed and used for integrating heterogeneous data from local or geographically distributed databases (20–22). However, integrating ‘omics’ data across different databases is still a challenge because of database heterogeneity, particularly the lack of a centralized vocabulary control for the metadata describing the experiment design, and the absence of unifying identifiers. A significant advantage of the RC is that all data has been integrated based on the UniProtKB accession number. These identifiers allow queries across data types and experiments, thereby enabling complex analyses of pathogen and host systems. By using the integrated data resource in the RC, researchers can be facilitated in their discovery and validation of pathogen and host interaction profiles.
Significance for systems biology and cyberinfrastructure
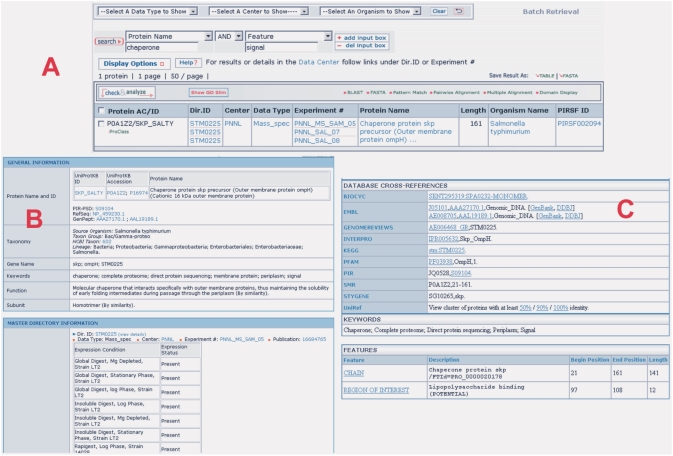
The advent of bioinformatics, genome-sequencing and high-throughput genome-wide experimentation (e.g. proteomics, transcriptomics) has lead to characterization of complex components pathosystems. System-wide studies of interactions between components of biological systems and how these interactions give rise to the function and behavior of that system are becoming increasingly possible (23–25). The available data in the RC [e.g. transcriptional and proteomics data of pathogen B. anthracis and of host mouse macrophages response (Use case 2)], greatly facilitates the analysis of the host and pathogen interaction using the framework of cyberinfrastructure built at the RC (26–30). For example, a researcher can query all proteins that have been experimentally demonstrated to interact with secretion system chaperones and further refine that list by choosing those proteins that have been annotated as having signal peptide characteristics and are conserved among a list of pathogens. This use case is illustrated in Figure 3. After entering the word ‘chaperone’ combined with the ‘protein name’ category, and ‘signal’ combined with the ‘feature’ category, as shown in the Figure 3A, and submitting the search, the system returns one chaperone protein in which the signal feature is represented (Figure 3A). Following the iProClass image (green at the left side), the user can review this chaperone protein summary information stored in the RC system (Figure 3B). Again clicking the UniProtKB ID hyperlink in Figure 3B, the user will obtain the most comprehensive annotation data regarding this chaperone protein (Figure 3C). More sophisticated search can be carried out by the experienced users.
Figure 3.
The studied Chaperone protein having a peptide signal feature. (A) search result by entering chaperone and signal keywords; (B) the summary information of Chaperone stored in the RC system; (C) the comprehensive annotation data of the chaperone protein.
Currently, several data sets including mass spectrometry, gene expression microarray, protein 3D structure and genomic clone data from several pathosystems are available for public access. As more data are integrated into the resource, it will become an even more valuable tool for the scientific community. We continue to improve the utility and usability of the resource to facilitate the research on the discovery of potential diagnostics, drug targets and vaccines.
FURTHER DEVELOPMENT
Experimental data sets continue to be submitted to the RC and are planned through June 2009. Ongoing development of the RC is driven by feedback from the PRC investigators, the scientific community and a Scientific Working Group http://www.proteomicsresource.org/AdminCenter/SWG.aspx for the project. We invite input from the research community through the Feedback form which can be reached from the top navigation bar on every RC page.
ACKNOWLEDGEMENTS
The authors appreciate comments and suggestions from Terry Brennan, Shamira Shallom, Joe Breen, Malu Polanski and JoJo Stemple. This work is funded through NIAID contract HHSN266200400061C. Funding to pay the Open Access publication charges for this article was provided by HHSN266200400061C.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ideker T, Winslow LR, Lauffenburger AD. Bioengineering and systems biology. Ann. Biomed. Eng. 2006;34:257–264. doi: 10.1007/s10439-005-9047-7. [DOI] [PubMed] [Google Scholar]
- 2.Smith JC, Figeys D. Proteomics technology in systems biology. Mol. Biosyst. 2006;2:364–370. doi: 10.1039/b606798k. [DOI] [PubMed] [Google Scholar]
- 3.de Hoog CL, Mann M. Proteomics. Annu. Rev. Genomics Hum. Genet. 2004;5:267–293. doi: 10.1146/annurev.genom.4.070802.110305. [DOI] [PubMed] [Google Scholar]
- 4.Waters KM, Pounds JG, Thrall BD. Data merging for integrated microarray and proteomic analysis. Brief Funct. Genomic Proteomic. 2006;5:261–272. doi: 10.1093/bfgp/ell019. [DOI] [PubMed] [Google Scholar]
- 5.Birkland A, Yona G. BIOZON: a system for unification, management and analysis of heterogeneous biological data. BMC Bioinformatics. 2006;7:70. doi: 10.1186/1471-2105-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng A, Bursteinas B, Gao Q, Mollison E, Zvelebil M. Resources for integrative systems biology: from data through databases to networks and dynamic system models. Brief Bioinform. 2006;7:318–330. doi: 10.1093/bib/bbl036. [DOI] [PubMed] [Google Scholar]
- 7.De Keersmaecker SC, Thijs IM, Vanderleyden J, Marchal K. Integration of omics data: how well does it work for bacteria? Mol. Microbiol. 2006;62:1239–1250. doi: 10.1111/j.1365-2958.2006.05453.x. [DOI] [PubMed] [Google Scholar]
- 8.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooksbank C, Cameron G, Thornton J. The European Bioinformatics Institute's data resources: towards systems biology. Nucleic Acids Res. 2005;33:D46–53. doi: 10.1093/nar/gki026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones P, Cote RG, Martens L, Quinn AF, Taylor CF, Derache W, Hermjakob H, Apweiler R. PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Res. 2006;34:D659–D663. doi: 10.1093/nar/gkj138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beavis RC. Using the global proteome machine for protein identification. Methods Mol. Biol. 2006;328:217–228. doi: 10.1385/1-59745-026-X:217. [DOI] [PubMed] [Google Scholar]
- 12.Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, et al. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–D658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leinonen R, Diez FG, Binns D, Fleischmann W, Lopez R, Apweiler R. UniProt archive. Bioinformatics. 2004;20:3236–3237. doi: 10.1093/bioinformatics/bth191. [DOI] [PubMed] [Google Scholar]
- 16.Wu CH, Huang H, Nikolskaya A, Hu Z, Barker WC. The iProClass integrated database for protein functional analysis. Comput. Biol. Chem. 2004;28:87–96. doi: 10.1016/j.compbiolchem.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Cammer S. SChiSM2: creating interactive web page annotations of molecular structure models using Jmol. Bioinformatics. 2007;23:383–384. doi: 10.1093/bioinformatics/btl603. [DOI] [PubMed] [Google Scholar]
- 18.Joyce AR, Palsson BO. The model organism as a system: integrating ‘omics’ data sets. Nat. Rev. Mol. Cell. Biol. 2006;7:198–210. doi: 10.1038/nrm1857. [DOI] [PubMed] [Google Scholar]
- 19.Cho CR, Labow M, Reinhardt M, van Oostrum J, Peitsch MC. The application of systems biology to drug discovery. Curr. Opin. Chem. Biol. 2006;10:294–302. doi: 10.1016/j.cbpa.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 20.Shannon PT, Reiss DJ, Bonneau R, Baliga NS. The Gaggle: an open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006;7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garwood K, Garwood C, Hedeler C, Griffiths T, Swainston N, Oliver SG, Paton NW. Model-driven user interfaces for bioinformatics data resources: regenerating the wheel as an alternative to reinventing it. BMC Bioinformatics. 2006;7:532. doi: 10.1186/1471-2105-7-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calder RB, Beems RB, van Steeg H, Mian IS, Lohman PH, Vijg J. MPHASYS: a mouse phenotype analysis system. BMC Bioinformatics. 2007;8:183. doi: 10.1186/1471-2105-8-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ideker T. Systems biology 101–what you need to know. Nat. Biotechnol. 2004;22:473–475. doi: 10.1038/nbt0404-473. [DOI] [PubMed] [Google Scholar]
- 24.Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 25.Werner E. All systems go. Nature. 2007;449:2. [Google Scholar]
- 26.Eckart JD, Sobral B.WS. A life scientist's gateway to distributed data management and computing: the PathPort/ToolBus Framework. OMICS: J. Integrative Biol. 2003;7:79–88. doi: 10.1089/153623103322006661. [DOI] [PubMed] [Google Scholar]
- 27.He YQ, Vines RR, Wattam AR, Abramochkin GV, Dickerman AW, Eckart JD, Sobral B.WS. PIML: The Pathogen Information Markup Language. Bioinformatics. 2005;21:116–121. doi: 10.1093/bioinformatics/bth462. [DOI] [PubMed] [Google Scholar]
- 28.Lathigra R, He Y, Vines R, Nordberg E, Sobral B. In: Genome Exploitation: Data Mining the Genome. Gustafson J, Shoemaker R, Snape JW, editors. New York, NY: Springer; 2005. pp. 183–196. [Google Scholar]
- 29.Snyder EE, Kampanya N, Lu J, Nordberg EK, Karur HR, Shukla M, Soneja J, Tian Y, Xue T, et al. PATRIC: The VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 2007;35:D401–D406. doi: 10.1093/nar/gkl858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobral BWS. Cyberinfrastructure for PathoSystems Biology. In. In: Setubal JC, Verjovski-Almeida S, editors. Advances in Bioinformatics and Computational Biology, Proceedings. Vol. 3594. Sao Leopoldo, Brazil: 2005. pp. 11–27. [Google Scholar]
- 31.Ratia K, Saikatendu KS, Santarsiero BD, Barretto N, Baker SC, Stevens RC, Mesecar AD. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl Acad. Sci. USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY, Walter JC, LaBaer J. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 33.Adkins JN, Mottaz HM, Norbeck AD, Gustin JK, Rue J, Clauss TR, Purvine SO, Rodland KD, Heffron F, et al. Analysis of the Salmonella typhimurium proteome through environmental response toward infectious conditions. Mol. Cell. Proteomics. 2006;5:1450–1461. doi: 10.1074/mcp.M600139-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Manes NP, Gustin JK, Rue J, Mottaz HM, Purvine SO, Norbeck AD, Monroe ME, Zimmer JS, Metz TO, et al. Targeted protein degradation by Salmonella under phagosome-mimicking culture conditions investigated using comparative peptidomics. Mol. Cell. Proteomics. 2007;6:717–727. doi: 10.1074/mcp.M600282-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Shi L, Adkins JN, Coleman JR, Schepmoes AA, Dohnkova A, Mottaz HM, Norbeck AD, Purvine SO, Manes NP, et al. Proteomic analysis of Salmonella enterica serovar typhimurium isolated from RAW 264.7 macrophages: identification of a novel protein that contributes to the replication of serovar typhimurium inside macrophages. J. Biol. Chem. 2006;281:29131–29140. doi: 10.1074/jbc.M604640200. [DOI] [PubMed] [Google Scholar]
- 36.Almeida MS, Johnson MA, Herrmann T, Geralt M, Wuthrich K. Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:3151–3161. doi: 10.1128/JVI.01939-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Brooun A, Griffith M, Moy K, Yadav MK, Velasquez J, et al. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J. Virol. 2006;80:7894–7901. doi: 10.1128/JVI.00467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Buchmeier MJ, Stevens RC, Kuhn P. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J. Virol. 2007;81:6700–6708. doi: 10.1128/JVI.02817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peti W, Johnson MA, Herrmann T, Neuman BW, Buchmeier MJ, Nelson M, Joseph J, Page R, Stevens RC, et al. Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7. J. Virol. 2005;79:12905–12913. doi: 10.1128/JVI.79.20.12905-12913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saikatendu KS, Joseph JS, Subramanian V, Clayton T, Griffith M, Moy K, Velasquez J, Neuman BW, Buchmeier MJ, et al. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1''-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saikatendu KS, Joseph JS, Subramanian V, Neuman BW, Buchmeier MJ, Stevens RC, Kuhn P. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J. Virol. 2007;81:3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergman NH, Anderson EC, Swenson EE, Janes BK, Fisher N, Niemeyer MM, Miyoshi AD, Hanna PC. Transcriptional profiling of Bacillus anthracis during infection of host macrophages. Infect. Immun. 2007;75:3434–3444. doi: 10.1128/IAI.01345-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergman NH, Anderson EC, Swenson EE, Niemeyer MM, Miyoshi AD, Hanna PC. Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J. Bacteriol. 2006;188:6092–6100. doi: 10.1128/JB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergman NH, Passalacqua KD, Gaspard R, Shetron-Rama LM, Quackenbush J, Hanna PC. Murine macrophage transcriptional responses to Bacillus anthracis infection and intoxication. Infect. Immun. 2005;73:1069–1080. doi: 10.1128/IAI.73.2.1069-1080.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]