Abstract
ORegAnno is an open-source, open-access database and literature curation system for community-based annotation of experimentally identified DNA regulatory regions, transcription factor binding sites and regulatory variants. The current release comprises 30 145 records curated from 922 publications and describing regulatory sequences for over 3853 genes and 465 transcription factors from 19 species. A new feature called the ‘publication queue’ allows users to input relevant papers from scientific literature as targets for annotation. The queue contains 4438 gene regulation papers entered by experts and another 54 351 identified by text-mining methods. Users can enter or ‘check out’ papers from the queue for manual curation using a series of user-friendly annotation pages. A typical record entry consists of species, sequence type, sequence, target gene, binding factor, experimental outcome and one or more lines of experimental evidence. An evidence ontology was developed to describe and categorize these experiments. Records are cross-referenced to Ensembl or Entrez gene identifiers, PubMed and dbSNP and can be visualized in the Ensembl or UCSC genome browsers. All data are freely available through search pages, XML data dumps or web services at: http://www.oreganno.org.
BACKGROUND
A consequence of the escalating pace of genomic sequencing has been the requirement for novel methodology and large-scale efforts to interpret and annotate sequence function. Initial efforts to achieve this were primarily focused on identifying protein-coding genes, RNA genes and repetitive DNA, since the rules governing their presence are generally tractable. However, less annotated, due to their small size and variability, gene regulatory sequences are widely regarded to be at least as important to our understanding of biological systems. To aid in their identification, computational techniques such as phylogenetic footprinting, transcription factor (TF)-binding matrices, and motif clustering have been developed (1–3). Unfortunately, the predictive ability of such methods has been difficult to assess without large, well-described and comprehensive collections of biologically validated regulatory sequences (3). Sets of cis-regulatory sequences have been annotated by curation from the primary literature and several databases have been developed to collect and disseminate these sets (4–11). However, these databases are often species- or process-specific, and do not provide sufficient details about the experiments or conditions under which function was demonstrated, and in some cases require payment for access. Data access is generally limited to web-based search pages without any option for the programmatic interaction essential to most bioinformatics studies. Finally, they are typically ‘closed systems’ in that they do not allow continued addition or annotation by the research community and as such are not maintainable over the long term without vast resources. We have developed the Open Regulatory Annotation database (ORegAnno) to overcome these challenges and support research in regulatory biology (12). ORegAnno provides standardized technologies for the long-term, community-driven, open-access curation of cis-regulatory data. Here we provide an update of developments on the ORegAnno database and progress in the field of open regulatory annotation.
OVERVIEW
ORegAnno (http://www.oreganno.org) is a database and literature curation system for community-based annotation of experimentally proven DNA regulatory regions, transcription factor binding sites (TFBS) and regulatory variants. A ‘publication queue’ allows papers of interest to be added to the system for future curation. Thus both regulatory papers and their regulatory sequences are managed in the system. ORegAnno is based on open-source technology and is comprised of a MySQL database with a Java-based web application that indexes new annotations using the Lucene search engine (http://lucene.apache.org/) and provides programmatic access to the underlying data using Hibernate (http://www.hibernate.org/) and SOAP Web Services. Figure 1 outlines the annotation process and information flow. Users in the gene regulation community can enter or ‘check out’ papers from the publication queue for detailed manual curation, using a series of annotation pages. A typical record entry consists of species, sequence type, sequence (plus sufficient flanking sequence for genome alignment), target gene, binding factor, experimental outcome and one or more detailed lines of experimental evidence demonstrating function of the sequence. Records are cross-referenced to Ensembl or Entrez Gene identifiers, PubMed and dbSNP (for regulatory polymorphisms). Before committing a record to the database, ORegAnno performs a number of error checks (e.g. that the sequence has not been entered previously) and asks the user to verify its contents. A BLAST-based mapping agent then assigns genome coordinates to each sequence, allowing it to be viewed as a track in the Ensembl or UCSC genome browsers. Once finished with a paper, a user will then ‘close’ it in the queue and assign an annotation result (success, neutral or failure). Existing records can be updated, commented and scored (positive if verified as correct; negative if a problem is identified) by any registered user or deprecated and replaced by a ‘Validator’ user. The complete database or any subset can be searched or downloaded in a number of formats or accessed programmatically.
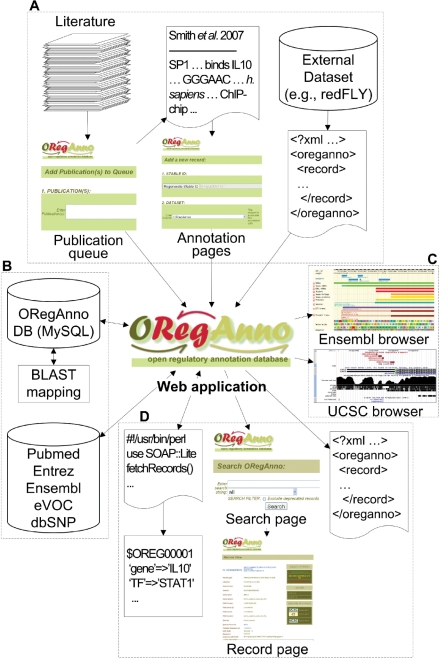
Figure 1.
Information flow for ORegAnno annotation process. (A) Data input. A publication queue allows papers from scientific literature to be added to the system for future curation. Users in the gene regulation community can enter or ‘check out’ papers from the queue for detailed manual curation using a series of user-friendly annotation pages. It is also possible to ‘batch upload’ complete datasets (e.g. external databases) using the ORegAnno XML data exchange format. (B) Data storage and processing. All functionality of the ORegAnno web application depends on storage and retrieval of data from an underlying MySQL relational database. Records are cross-referenced to PubMed, Entrez, Ensembl, dbSNP and eVOC where appropriate. A BLAST-based mapping agent assigns genome coordinates to each sequence. (C) Visualization. All mapped ORegAnno records can be viewed as custom tracks in the Ensembl or UCSC genome browsers. Most records are also available as official tracks in UCSC. (D) Data access. The web application provides an advanced search page for the entire record set. Each record page represents a complete summary of the data for a verified regulatory sequence. Nightly data dumps are posted in XML format. Programmatic interaction with ORegAnno is available through web services using the Perl SOAP modules.
RECENT DEVELOPMENTS
New entries
Since ORegAnno was first released, the collection has grown by ∼10-fold from 2691 to 30 145 records. This total includes 15 738 regulatory regions, 14 229 TFBSs and 178 regulatory variants (polymorphisms and haplotypes) from 19 species (Table 1). A total of 29 433 records have been mapped to one of 14 species representing a mapping success rate of ∼98%. New additions were incorporated from external datasets including a large set of human promoters (13), the REDfly resource (9), HBB and Erythroid modules (14,15), the Vista Enhancer dataset (11), ChIP–chip sites for CTCF(16) and multiple yeast TFs(17,18) and ChIP-Seq sites for STAT1 (19) and REST (20). Apart from the 11 external datasets currently in ORegAnno, extensive manual curation of the literature has produced an additional 1293 original sequence records. A large number of annotations were entered during the RegCreative Jamboree (http://www.dmbr.ugent.be/bioit/contents/regcreative/) at which 130 scientific articles were examined in depth with 96 papers meeting the criteria for annotation and resulting in 501 new regulatory sequence records. In total, 922 publications have been curated by 45 contributing users (from >300 registered users). The complete set of records contain regulatory sequences for over 3853 genes and 465 TFs, describe 41 856 experimental sources of evidence referencing 31 different cell types and are further annotated by 49 807 user-comments. The majority of records (98.9%) had positive experimental outcomes (i.e. the experiments demonstrated the sequence to be functional) but a small set of negative or neutral results have also been catalogued.
Table 1.
Current content of ORegAnno database
| Species | Regulatory haplotype | Regulatory polymorphism | Regulatory region | Transcription factor binding site | Totals |
|---|---|---|---|---|---|
| Bos taurus | 1 | 1 | |||
| Caenorhabditis briggsae | 21 | 21 | |||
| Caenorhabditis elegans | 13 | 194 | 207 | ||
| Ciona intestinalis | 7 | 17 | 24 | ||
| Ciona savignyi | 1 | 1 | 2 | ||
| Cricetinae | 3 | 3 | |||
| Danio rerio | 2 | 2 | 4 | ||
| Drosophila melanogaster | 680 | 1415 | 2095 | ||
| Gallus gallus | 8 | 29 | 37 | ||
| Halocynthia roretzi | 6 | 6 | |||
| Homo sapiens | 6 | 171 | 14 948 | 7834 | 22 959 |
| HIV 1 | 2 | 2 | |||
| Mus musculus | 1 | 55 | 215 | 271 | |
| Oryctolagus cuniculus | 1 | 1 | |||
| Rattus norvegicus | 15 | 99 | 114 | ||
| Saccharomyces cerevisiae | 1 | 4392 | 4393 | ||
| Takifugu rubripes | 2 | 2 | |||
| Xenopus laevis | 1 | 1 | 2 | ||
| Xenopus tropicalis | 1 | 1 | |||
| Totals (19 species) | 7 | 171 | 15 738 | 14 229 | 30 145 |
Recent applications
The ORegAnno resource has proven useful for the development of both computational and experimental methods for the identification of novel TFBSs and regulatory polymorphisms. One such approach, called cisRED (http://www.cisred.org), uses multiple motif discovery methods applied to sequence sets that include up to 42 orthologous sequence regions from vertebrates (21). The collection of known binding sites in ORegAnno has proved an invaluable resource for the parameter optimization and estimates of accuracy for this resource. In another study, the set of known regulatory SNPs (rSNPs) in ORegAnno was used to investigate and prioritize various properties that may be important for identifying novel regulatory polymorphisms (22). The discriminatory potential of 23 properties related to gene regulation and population genetics was assessed by comparing these known rSNPs to a set of SNPs of unknown function (ufSNPs). A support vector machine classifier using these properties was able to discriminate rSNPs from ufSNPs with a sensitivity and specificity of 82% and 71%, respectively (22). Finally, ORegAnno has also served a critical role in the development of new experimental approaches such as ChIP-Seq. ChIP-Seq is similar to the well-described ChIP–chip method (23) except that DNA fragments isolated from the protein–DNA complex are identified by DNA sequencing instead of hybridization to a tiling microarray. The approach was first demonstrated for the STAT1 TF in interferon-γ-stimulated HeLa S3 cells (19). A set of 41 experimentally verified sites representing 34 genomic loci for STAT1 binding were first collected from the literature and entered into ORegAnno (Oreganno dataset: OREGDS00006). Stimulated ChIP-Seq peaks were found to overlap 24 of 34 of these loci, suggesting a sensitivity of ∼71%. For the ORegAnno STAT1 sites shown to be functional in HeLa cells specifically, sensitivity was 100%. The collection of known STAT1 sites and binding matrices derived from them also allowed a set of high-confidence novel STAT1-binding sites to be determined and entered into ORegAnno as their own dataset (OREGDS00007). This iterative process, whereby existing data drives the creation of new data, demonstrates the utility and flexibility of the ORegAnno system.
Publication queue
An important new feature of ORegAnno called the ‘publication queue’ was created as a literature management system to allow registered users to input relevant papers from the scientific literature as targets for annotation. All that is required to enter a publication is a valid PubMed identifier. Optionally, a TF can be specified, allowing users to later search the queue for papers related to TFs of interest. Normally, publications are added to the queue with an entry type of ‘expert entry’, indicating that a human expert reviewed the paper and found it to be relevant. However, it is also possible to enter ‘text-mining entry’ papers (see below). A publication enters the queue with an initial state of ‘pending’. Any user can then ‘open’ the publication and begin the annotation process. Once annotated, the paper is either ‘closed’ or reset to ‘pending’ if annotation work remains. Free-form comment fields are optional for each change of state. However, when a publication is closed, one of several standardized closure comments must be chosen (success – addition of new records, failure – did not describe regulatory element, etc.). These allow the overall success rate and failure causes to be tracked. The queue can be queried on a number of fields including user, PubMed id, title, abstract, author, publication date and journal. Search results can be optionally filtered by state (pending, open or closed), TF, entry type (expert or text mining) or text-mining score. Each queue record contains a history of all state changes and comments as well as links to the publication's PubMed abstract. The current set of ‘expert entry’ papers in the queue was obtained from existing sources of curated publications including the Drosophila DNase I Footprint Database (8), REDfly (9), a catalog of regulatory elements for muscle-specific regulation of transcription (24,25), ABS (4), TRED (7), ooTFD (26), DBTGR (10) or added manually by individual ORegAnno users from literature searches and review articles. The expert entry queue currently contains 4438 gene regulation papers of which 3478 are open or pending and 960 are closed.
Development of text-mining strategies and the ‘text-mining queue’
The publication queue represents an unprecedented resource for researchers interested in developing text-mining approaches to identify papers involved in gene regulation and/or extract regulatory data from these papers. We used both the ‘success’ and the ‘failure’ papers from the ‘expert-entry’ queue to validate and compare different vector space models (27) for cis-regulatory document retrieval (Aerts and coworkers, manuscript in preparation). The model with the best performance in terms of sensitivity and specificity was chosen to rank the entire corpus of PubMed abstracts. By manually curating uniformly distributed samples from the top 100 000 scoring abstracts, a cut-off was set at ∼58 000 so that the positive predictive value (PPV) of top-scoring abstracts reached 50%, a success rate similar to that achieved during the RegCreative Jamboree (54%), and judged satisfactory by the Jamboree participants. These 58 000 papers, containing an estimated 29 000 papers that will result in regulatory annotations, have been added to the ORegAnno queue (54 351 new additions after removing duplications). We estimate that this large cis-regulatory text corpus will require around 15–30 person-years to be fully curated. Therefore, the Open Regulatory Annotation Consortium is actively pursuing research in text-mining techniques to identify the actual cis-regulatory sequences, the species and the target gene automatically from the full text papers. In a pilot study, sequences were extracted from full-text articles for papers in the ORegAnno expert-based queue and the top 4501 papers from the text-mining-based queue. When comparing the automatically extracted data with the collection of manual ORegAnno annotations, this study achieved a reasonably high PPV (62%) at the sequence level, showing that automatic draft annotation of cis-regulatory elements is indeed feasible by text-mining (Aerts and coworkers, manuscript in preparation). Such draft annotations should help accelerate the manual curation and can also serve directly as benchmark data to validate cis-element prediction algorithms.
Ontologies in ORegAnno
The ORegAnno evidence ontology is a simple ontology of evidence classes, types and subtypes for describing experiments that demonstrate the identity and/or function of regulatory sequences and their factors. These lines of evidence capture critical details from primary experiments and allow end users to filter the ORegAnno sequence set, based on their own criteria for experimental credibility. The ontology has been considerably extended since last published, and currently consists of six classes (e.g. Transcription regulator site), 14 evidence types (e.g. Reporter gene assay) and 72 evidence subtypes (e.g. Transient transfection luciferase assay). This ontology has been adopted by the PAZAR resource (28) and is being developed in collaboration with that group using Protégé (http://protege.stanford.edu/). The complete evidence ontology can be obtained in XML format (http://www.oreganno.org/oregano/evidence.xml) or as a Protégé project file (http://www.pazar.info/ontologies/newevidence.pprj). Within each line of evidence, a user can also specify the cell type in which experiments were conducted using the eVOC cell type ontology (29). We are working to incorporate additional Ontologies such as the BRENDA Tissue Ontology, and improvements to the Sequence Ontology are currently being developed for the cis-regulatory domain.
Other improvements
The ORegAnno website has been updated to use Ajax technology, improving the ease of annotation. Ajax improves a web page's usability by exchanging small amounts of data with the server behind the scenes, so that the entire web page does not have to be reloaded each time the user requests a change (http://www.xul.fr/en-xml-ajax.html). A detailed case study has been added to the help pages to guide users through the entire process of annotating a paper. Annotation pages have been improved so that individual ‘help bubbles’ are available next to each field. Additional web services methods have been created to allow programmatic access to the publication queue and genome mappings.
DATA ACCESS
The website (http://www.oreganno.org) provides access to an advanced search page for the entire record set, the publication queue, simple tools for scanning or extracting sequences, database dumps and extensive help documentation. Each record page represents a complete summary of the data for a verified regulatory sequence along with links to external sources such as UCSC, Ensembl and PubMed. All data are freely available in a number of formats without any user registration. Users are required to register and login only if they wish to add records, comments or scores. Nightly data dumps of the database are posted in XML format on the website. Human (hg18) and fly (dm3) records are available through the UCSC genome browser (http://genome.ucsc.edu/) as a standard track under the ‘Expression and Regulation’ tab. Mouse (mm8), worm (ce4) and rat (rn4) are available through the UCSC ‘genome-test’ browser (http://genome-test.cse.ucsc.edu/). The ORegAnno dataset is also in the process of being incorporated into the PAZAR database (760 records to date). Programmatic interaction with ORegAnno is available through web services using the Perl SOAP modules (see ‘Dump’ page for examples). Requests for the entire database (e.g. a MySQL dump) or other formats can be addressed to the authors. ORegAnno records are typically mapped to only the most current genome build for each species as provided by UCSC (e.g. hg18 for human). However, mapping can easily be performed for any other genome build upon request. A mailing list exists for updates and user assistance (oreganno@bcgsc.ca). The ORegAnno web application is available open-source under the Lesser GNU Public License at https://oreganno.dev.java.net/.
ACKNOWLEDGEMENTS
We thank the Open Regulatory Annotation Consortium for their continuing efforts to improve this resource through manual curation and record validation. We also thank the owners of regulatory sequence databases that made their data available for inclusion in ORegAnno. This work was funded by British Columbia Cancer Foundation; Genome Canada; Genome British Columbia; European Network of Excellence (ENFIN); BioSapiens Network of Excellence; Research Foundation – Flanders (FWO); The Pleiades Promoter Project; Michael Smith Foundation for Health Research to O.L.G., M.C.S., M.G. and S.J.M.J.; Canadian Institutes of Health Research to O.L.G.; European Molecular Biology Laboratory to S.B.M.; Marie Curie Early Stage Research Training Fellowship (MEST-CT-2004-504854) to M.H.; Natural Sciences and Engineering Research Council to S.B.M., and M.G.; Research Foundation – Flanders (FWO) to P.V.L.; Swedish Research Council to C.W. Funding to pay the Open Access publication charges for this article was provided by Genome Canada and Genome British Columbia.
Conflict of interest statement. None declared.
THE OPEN REGULATORY ANNOTATION CONSORTIUM MEMBERS
Amy Ticoll, Andy Schroeder, Arun Ramani, Bart Hooghe, Belinda Giardine, Boris Adryan, Bridget Bernier, Casey Bergman, Claes Wadelius, Daniel Sobral, Debra Fulton, Denis Thieffry, Dominique Vlieghe, Elodie Portales-Casamar, Enrique Blanco, Erin D. Pleasance, Florian Leitner, Gordon Robertson, Hedi Peterson, Helge Roider, Ian J. Donaldson, Ildefonso Cases, Jean Imbert, Jean-Valery Turatsinze, Jonathan Mudge, Katayoon Kasaian, Maggie Zhang, Malachi Griffith, Marc Halfon, Maximilian Haeussler, Misha Bilenky, Monica Sleumer, Nathalie Theret, Nikiforos Karamanis, Obi Griffith, Paco Hulpiau, Peter Van Loo, Pieter De Bleser, Priit Adler, Ross Hardison, Shaun Mahony, Stein Aerts, Stephen Montgomery, Steven J.M. Jones, Steven M. Gallo, Wyeth Wasserman, Yves Moreau.
REFERENCES
- 1.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 2.Elnitski L, Jin VX, Farnham PJ, Jones SJ. Locating mammalian transcription factor binding sites: a survey of computational and experimental techniques. Genome Res. 2006;16:1455–1464. doi: 10.1101/gr.4140006. [DOI] [PubMed] [Google Scholar]
- 3.Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 4.Blanco E, Farre D, Alba MM, Messeguer X, Guigo R. ABS: a database of Annotated regulatory Binding Sites from orthologous promoters. Nucleic Acids Res. 2006;34:D63–D67. doi: 10.1093/nar/gkj116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang C, Xuan Z, Zhao F, Zhang MQ. TRED: a transcriptional regulatory element database, new entries and other development. Nucleic Acids Res. 2007;35:D137–D140. doi: 10.1093/nar/gkl1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergman CM, Carlson JW, Celniker SE. Drosophila DNase I footprint database: a systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics. 2005;21:1747–1749. doi: 10.1093/bioinformatics/bti173. [DOI] [PubMed] [Google Scholar]
- 9.Gallo SM, Li L, Hu Z, Halfon MS. REDfly: a regulatory element database for Drosophila. Bioinformatics. 2006;22:381–383. doi: 10.1093/bioinformatics/bti794. [DOI] [PubMed] [Google Scholar]
- 10.Sierro N, Kusakabe T, Park KJ, Yamashita R, Kinoshita K, Nakai K. DBTGR: a database of tunicate promoters and their regulatory elements. Nucleic Acids Res. 2006;34:D552–D555. doi: 10.1093/nar/gkj064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montgomery SB, Griffith OL, Sleumer MC, Bergman CM, Bilenky M, Pleasance ED, Prychyna Y, Zhang X, Jones SJ. ORegAnno: an open access database and curation system for literature-derived promoters, transcription factor binding sites and regulatory variation. Bioinformatics. 2006;22:637–640. doi: 10.1093/bioinformatics/btk027. [DOI] [PubMed] [Google Scholar]
- 13.Trinklein ND, Aldred SJ, Saldanha AJ, Myers RM. Identification and functional analysis of human transcriptional promoters. Genome Res. 2003;13:308–312. doi: 10.1101/gr.794803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Zhang Y, Cheng Y, Zhou Y, King DC, Taylor J, Chiaromonte F, Kasturi J, Petrykowska H, et al. Experimental validation of predicted mammalian erythroid cis-regulatory modules. Genome Res. 2006;16:1480–1492. doi: 10.1101/gr.5353806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 20.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 21.Robertson G, Bilenky M, Lin K, He A, Yuen W, Dagpinar M, Varhol R, Teague K, Griffith OL, et al. cisRED: a database system for genome-scale computational discovery of regulatory elements. Nucleic Acids Res. 2006;34:D68–D73. doi: 10.1093/nar/gkj075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montgomery SB, Griffith OL, Schuetz JM, Brooks-Wilson A, Jones SJ. A survey of genomic properties for the detection of regulatory polymorphisms. PLoS Comput. Biol. 2007;3:e106. doi: 10.1371/journal.pcbi.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 24.Wasserman WW, Fickett JW. Identification of regulatory regions which confer muscle-specific gene expression. J. Mol. Biol. 1998;278:167–181. doi: 10.1006/jmbi.1998.1700. [DOI] [PubMed] [Google Scholar]
- 25.Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh D. Object-oriented transcription factors database (ooTFD) Nucleic Acids Res. 2000;28:308–310. doi: 10.1093/nar/28.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glenisson P, Coessens B, Van Vooren S, Mathys J, Moreau Y, De Moor B. TXTGate: profiling gene groups with text-based information. Genome Biol. 2004;5:R43. doi: 10.1186/gb-2004-5-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Portales-Casamar E, Kirov S, Lim J, Lithwick S, Swanson MI, Ticoll A, Snoddy J, Wasserman WW. PAZAR: a framework for collection and dissemination of cis-regulatory sequence annotation. Genome Biol. 2007;8:R207. doi: 10.1186/gb-2007-8-10-r207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelso J, Visagie J, Theiler G, Christoffels A, Bardien S, Smedley D, Otgaar D, Greyling G, Jongeneel CV, et al. eVOC: a controlled vocabulary for unifying gene expression data. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]