Abstract
Embryonic exposure to the endocrine disruptor vinclozolin during gonadal sex determination appears to promote an epigenetic reprogramming of the male germ-line that is associated with transgenerational adult onset disease states. Transgenerational effects on the embryonic day 16 (E16) testis demonstrated reproducible changes in the testis transcriptome for multiple generations (F1-F3). The expression of 196 genes were found to be influenced, with the majority of gene expression being decreased or silenced. Dramatic changes in the gene expression of methyltransferases during gonadal sex determination were observed in the F1 and F2 vinclozolin generation (E16) embryonic testis, but the majority returned to control generation levels by the F3 generation. The most dramatic effects were on the germ-line associated Dnmt3A and Dnmt3L isoforms. Observations demonstrate that an embryonic exposure to vinclozolin appears to promote an epigenetic reprogramming of the male germ-line that correlates with transgenerational alterations in the testis transcriptome in subsequent generations.
Keywords: Transgenerational, Epigenetics, DNA Methylation, Testis, Transcriptome, Microarray, Endocrine Disruptor, Environmental Toxicology, Sex Determination, Embryonic Development, Vinclozolin
INTRODUCTION
Although a large number of different genes have been shown to be associated with a variety of different diseases, the majority do not have DNA sequence mutations that can explain altered gene expression. In addition, a number of environmental factors and compounds have been shown to influence a variety of diseases [1], but few have been shown to promote DNA sequence mutations, suggesting an alternate mechanism not involving changes in DNA sequence [2]. Epigenetics can influence the gene expression profiles and transcriptomes of most organs and cell types [3]. Therefore, alterations in the epigenome appears to be a major factor in the regulation of the transcriptomes associated with disease [4]. Epigenetics is an important mechanism in the ability of environmental factors to influence health and disease [5].
Previous observations have demonstrated that the embryonic exposure to endocrine disruptors (i.e. vinclozolin and methoxychlor) during gonadal sex determination induces a transgenerational effect on adult male reproduction and sperm production [6]. Recently, an extension of this study demonstrated that as vinclozolin exposed generation animals age (i.e. 6–14 months) a variety of different disease states develop in a transgenerational manner (i.e. F1-F4), including breast tumors, prostate disease, kidney disease and immune abnormalities [7]. Therefore, exposure to the endocrine disruptor vinclozolin during gonadal sex determination promoted the development of a variety of different adult onset diseases [7; 8]. The transgenerational phenotype was transmitted through the male germ-line [6; 7]. Although females did develop disease [7], they did not transmit this phenotype to the next generation [6; 7]. The potential contribution of the female germ-line to the transgenerational phenotype remains to be elucidated. Alterations in the male germ-line epigenome was identified following endocrine disruptor (i.e. vinclozolin) exposure [6]. This epigenetic reprogramming of the male germ-line appears to allow the disease phenotype to become transgenerational [6; 7; 8].
Endocrine disruptors are compounds that bind hormone receptors or effect hormone signaling to alter normal endocrinology [9]. A large number of environmental compounds, from plastics to pesticides, have endocrine disruptor activity [9]. The endocrine disruptor used in the current study was vinclozolin, which is a fungicide used in the fruit industry [10]. Vinclozolin and its metabolites are anti-androgenic compounds that bind and alter androgen receptor actions [11]. A number of studies have shown that embryonic and early postnatal exposure to endocrine disruptors can cause subsequent adult onset diseases [10; 11; 12]. The potential that endocrine disruptor actions are mediated in part through alterations of the epigenome that are causal in the mechanism of disease remains to be elucidated.
The biological process involved in the transgenerational epigenetic disease phenotype involves the epigenetic programming of the germ cells during gonadal sex determination [13; 14]. As the primordial germ cells migrate to the genital ridge and colonize the bipotential gonad their DNA becomes de-methylated [13; 14]. The germ cell DNA is then re-methylated after gonadal sex determination and during gonadal differentiation in a sex specific manner [15]. Therefore, the critical period to influence the germ-line DNA methylation is during this epigenetic programming of the germ cells [6; 15]. The ability to induce permanent alterations in the germ-line DNA methylation pattern is hypothesized to in part allow the phenotype to become transgenerational [6; 7]. The influence this altered germ-line epigenome subsequently has on the transcriptomes of developing organs was investigated in the current study using the embryonic testis as a model organ. Observations demonstrate a transgenerational effect of the endocrine disruptor vinclozolin on the embryonic testis transcriptome.
RESULTS
Endocrine Disruptor Actions
Gestating rats were exposed to a daily intraperitoneal (IP) injection of vinclozolin during E8-E14 of embryonic development corresponding to the period of gonadal sex determination [6; 12]. Litter mate sisters were used for the control (vehicle) and vinclozolin treatments. Eight different gestating mothers were used for each control and vinclozolin treatment population. The F1 vinclozolin generation males were bred to F1 vinclozolin generational females from different litters to generate the F2 vinclozolin generation, and then F2 generation animals were bred to generate the F3 generation. Both the control and vinclozolin generations were bred in a similar manner. No sibling breeding was done and control and vinclozolin generation animals were housed, fed and maintained under similar conditions. The embryonic day 16 (E16) testes were collected for histological analysis and RNA preparation. The morphology of the E16 testis was similar between the control and vinclozolin F1-F3 generation animals, Figure 1. The analysis of cellular apoptosis demonstrated an increase in germ cell death between the control and vinclozolin F1 and F2 generation E16 testes, but not F3, Figure 1E. A wildtype E16 testis level of apoptosis is also shown for comparison. The effects on the F1 and F2 E16 testis cell apoptosis are likely due in part to direct exposure and toxicology of the F1 embryo and F2 generation germ-line. The F3 generation is the first generation not directly exposed, so is the first generation that is unequivocally transgenerational (i.e. transgenerational generation). Although morphological abnormalities develop in the testis of adult animals [6], no major effects were observed during embryonic development, Figure 1.
Figure 1.
Embryonic day 16 (E16) testis histology and cellular apoptosis. E16 testis from F2 control (A, C) and vinclozolin (B, D) generation animals for histology (A, B) and TUNEL apoptosis staining (C, D). The number of apoptotic germ cells/section (E) for F1-F3 control and vinclozolin generation E16 testes are presented, mean ± SEM, from three different experiments, and compared to control wildtype rat E16 testis (Wildtype). An asterisk (*) indicates a statistical difference from control (p<0.05).
Transgenerational Transcriptome Regulation
The transgenerational changes in the testis transcriptome were investigated with a microarray analysis of the E16 testis. The RNA from three different litters were pooled (i.e. 18–25 male pups) for an individual microarray chip and experiment. Two different experiments (i.e. microarray chips and animal sets) were compared for control and vinclozolin F1-F3 generations. Initially, an older rat Affymetrix RG-U34A chip was used that had 8,000 genes and similar results to those described below were obtained (data not shown). A more recent Affymetrix R230 2.0 gene chip was used that contained a larger percentage of the genome, 30,000 transcripts, and RNA from a different set of experiments and animals. All the replicate microarray chips were highly reproducible with an R2 >0.98. Genes were identified with a raw statistically significant (p<0.05) present call signal greater than 75 and a greater than 1.5-fold change in expression between control and vinclozolin generation E16 testis samples. A dendrogram analysis is shown in Figure 2A and demonstrates the control F1-F3 microarrays are essentially the same with negligible differences. Compared to controls, the F1 vinclozolin E16 testis had 2,071 genes altered, the F2 vinclozolin had 1,375 genes altered and the F3 vinclozolin E16 testis had 566 genes altered, Figure 3A. The majority (90%) of genes altered had a decrease in expression, Figure 2A, with approximately 10% with an increase in expression. A comparison of F1-F3 vinclozolin generation E16 testis transcriptome changes demonstrated 196 genes had similar altered expression in all generations, Figure 3A. Therefore, the vinclozolin F1-F3 generation animals had similar changes in the E16 testis transcriptome for these 196 genes, Figure 2B, and most were decreased in comparison to the control generations, Figure 2C. Due to the potential toxicology of vinclozolin in the F1 generation animals it is not surprising that a larger number of genes were affected (i.e. 2,071). Since the F2 generation germ-line was also directly exposed to vinclozolin, the number of affected genes in the F2 generation is also high (i.e. 1,375). Although the F2 generation could involve transgenerational effects, the F3 generation is the first generation that has had no direct exposure, so it is unequivocally transgenerational. Therefore, the F3 generation genes have a transgenerational change in expression. A preliminary analysis of the F4 generation E16 testis transcriptome was performed, however, the arrays were performed at different times than the F1-F3 arrays limiting direct comparison (data not shown). This F4 generation experiment would determine and potentially further decline in transgenerational effects.
Figure 2.
The E16 testis transcriptome microarray analysis from F1-F3 control and vinclozolin generation animals. (A) Dendogram for total regulated genes with signal above 75. (B) Dendogram of 196 regulated gene list. (C) The 196 gene list relative expression levels between F1-F3 controls and vinclozolin (vincl) E16 testis. The scale in the right margin indicates no change (black), increase (red) and decrease (green).
Figure 3.
Categorization of the F1-F3 regulated genes. (A) Venn diagram with total regulated (>1.5 fold-change between control and vinclozolin) genes listed and the overlap, with 196 shown to be consistent between the F1-F3 generations. (B) Categorization into functionally related gene groups with the number of genes (blue down-regulated and yellow up-regulated).
Previous observations suggested the transgenerational disease phenotype was transmitted through the paternal line [6; 7]. A vinclozolin outcross (VOC) experiment with an F2 vinclozolin generation male and a wildtype female was generated for a microarray analysis of the F3 VOC E16 testis. Comparison of the altered transcriptomes demonstrated the F3 VOC had similar alterations as the F1-F3 vinclozolin E16 testis with 165 genes of the 196 gene list being similar, Supplemental Table S1. Therefore, as observed with the adult disease phenotype [6; 7], the transgenerational alterations in the transcriptome appear to be transmitted primarily through the paternal line. Potential contributions of the female germ-line remain to be elucidated and no experiments of the embryonic female gonad transcriptome has been performed.
The 196 genes that were altered transgenerationally in the F1, F2, and F3 vinclozolin generation were further analyzed. Cluster analysis of categories of genes demonstrated that genes involved in transcriptional regulation, signal transduction, and cytoskeleton were highly represented, Figure 3B. Other gene categories represented included metabolism, cell cycle, development, proteolysis and apoptosis. Expressed sequence tags and unknown genes were also highly represented. The majority (90%) of genes were down-regulated in the vinclozolin generation (F1-F3) E16 testis, with 10% up-regulated, Figures 2 and 3B. The full list of 196 genes and relative signals is presented in Supplemental Table S1. A subset of genes (i.e. 90) from the 196 gene list that were significantly down-regulated and/or silenced in the F1-F3 vinclozolin generation E16 testis is shown in Supplemental Table S2. These genes provide candidates for direct or indirect epigenetic modification that may silence gene expression.
Sites of Potential Epigenetic Regulation
DNA methylation patterns are established during embryogenesis through cooperation of a family of methyltransferases (Dnmts) that include Dnmt1, Dnmt2, Dnmt3A, Dnmt3B, and Dnmt3L [16; 17; 18]. There are also Dnmt associated proteins (e.g. Dmap1) and methyltransferases for histones (e.g. euchromatic histone methyltransferase, Ehmt1) and RNA (e.g. RNA methyltransferase, Rnmt, and tRNA-methyltransferase 1, Trmt1). These methyltransferases in part regulate the epigenome of developing organs and the germ-line. The expression of several of these methyltransferases in control and vinclozolin F1-F3 generation E16 testis is shown in Figure 4. All the genes had a statistically significant change in expression in the F1 or F2 vinclozolin generation E16 testis compared to control, Table 1. The DNA methyltransferases (Dnmts) and histone methyltransferase (Ehmt1) all decreased, with the most dramatic change in Dnmt3A, Figure 4. Dnmt3A has been shown to be essential in paternal and maternal imprinting of the germ-line [19]. The Dmap1, Trmt1 and Rnmt1 all increased in expression in the vinclozolin samples. Therefore, the alteration in the epigenome induced by vinclozolin may be in part due to alterations in methyltransferase activity during F1 generation embryonic exposure. Interestingly, the F3 generations E16 testis gene expression levels appear to be returning to the normal control levels, except Ehmt1, Figure 4. Therefore, the initial effects may be in part mediated through alterations in normal methyltransferase activity, but the transgenerational transmission appears to be through an alternate mechanism, such as a modified epigenome in the germ-line.
Figure 4.
Methyltransferase gene expression in the F1-F3 control and vinclozolin generation E16 testis. Relative expression is presented for the specific genes after microarray analysis. The F1 and F2 vinclozolin generation gene expression for Dnmt3A, Dmap1, Dnmt2, Ehmt1 and Rnmt values were statistically different from the respective control generation values (p<0.05), with Dnmt1 statistically different in the F2 vinclozolin generation samples and Trmt1 not statistically different in any generation.
Table 1.
| A. F1 Generation Regulated Epigenetic Associated Genes | ||||
|---|---|---|---|---|
| Microarray Signal | ||||
| Gene Category & Symbol | Control | Vinclozolin | Genbank # | Description/Function |
| Methylation | ||||
|
| ||||
| Dnmt3A | 110 | 44 | AA956455 | DNA methyltransferase 3A |
| Dnmt2 | 68 | 47 | AI17081 | DNA methyltransferase 2 |
| Hrmt113 | 42 | 82 | AF059530 | Heterogeneous nuclear ribonucleoprotin methlytransferrase-like 3 |
| Gadd45A | 79 | 52 | NM_024127 | Growth arrest and DNA-damage inducible 45 alpha |
|
| ||||
| Histone Modification | ||||
|
| ||||
| Mgea5 | 323 | 185 | BF548107 | Meningioma expressed antigen 5 (hyaluronidase) |
| RGD1311017_pre | 82 | 35 | B1289182 | Similar to multiple hat domains (predicted) |
| Hdac1 | 563 | 200 | AW530195 | Histone deacetylase 1 (predicted) |
| Ash1l_pre | 144 | 57 | BG663056 | Ash1 (absent, small or homeotic)-like (predicted) |
| Ehmt1_pre | 496 | 321 | BM389055 | Euchromatic histone methyltransferase 1 (predicted) |
| LOC679252 | 235 | 141 | A1070638 | Similar to (Histone-lysine N-methlytransferase, H3) |
|
| ||||
| Chromatin Remodeling | ||||
|
| ||||
| Rere | 75 | 35 | NM_053885 | Arginine-glutamic acid dipeptide (RE) repeats |
| Mta1 | 272 | 157 | AJ132046 | Metastasis associated 1 |
| Smarca5_l | 215 | 139 | BF557855 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 (predicted) |
| Chd1_pre | 378 | 115 | BF396625 | Chromodomain helicase DNA binding protein 1 (predicted) |
| Chd2_pre | 178 | 95 | BF396633 | Chromodomain helicase DNA binding protein 2 (predicted) |
| Chd6_pre | 57 | 93 | BF396625 | Chromodomain helicase DNA binding protein 6 (predicted) |
| Shprh_pre | 97 | 60 | BE104039 | SNF2 hisotne linker PHD RING helicase (predicted) |
|
| ||||
| Transcription Regulation | ||||
|
| ||||
| Epc2_pre | 233 | 113 | AW918173 | Enhancer of polycomb homolog 2 (predicted) |
| Pcgf6 | 419 | 256 | AA858786 | Polycomb group ring finger 6 |
| B. F2 Generation Regulated Epigenetic Associated Genes | ||||
| Microarray Signal | ||||
| Gene Category & Symbol | Control | Vinclozolin | Genbank # | Description/Function |
| Methylation | ||||
|
| ||||
| Dnmt3A | 127 | 56 | AA956455 | DNA Methyltransferase 2 |
| Dnmt1 | 558 | 417 | AI179516 | DNA (cytosine-5)-methyltransferase 1 |
| Tpmt | 138 | 69 | BG381002 | leucine carboxyl methyltransferase 1 |
|
| ||||
| Histone Modification | ||||
|
| ||||
| Ash1l_pre | 139 | 54 | BG663056 | ash1 (absent, small, or homeotic)-like (predicted) |
| Hdac1 | 532 | 279 | AW530195 | histone deacetylase 1 |
| Ehmt1 | 493 | 333 | BM3890 | Eucharomatic histone methylatransferase 1 |
| Jmjd3_pre | 360 | 225 | BE118720 | jumonji domain containing 3 (predicted) |
| RGD1311017_pre | 91 | 41 | BI289182 | similar to multiple hat domains (predicted) |
| LOC679252 | 236 | 129 | AI070638 | similar to (Histone-lysine N-methyltransferase, H3) |
| RGD1566399_pre | 273 | 174 | BE098769 | Similar to MYST histone acetyltransferase monocytic leukemia 4 (predicted) |
|
| ||||
| Chromatin Remodeling | ||||
|
| ||||
| Cbx1_pre | 330 | 98 | BF389675 | chromobox homolog 1 (Drosophila HP1 beta) (predicted) |
| Chd1_pre | 386 | 158 | BF396625 | chromodomain helicase DNA binding protein 1 (predicted) |
| Chd4 | 173 | 86 | BF412612 | chromodomain helicase DNA binding protein 4 |
| RGD1561537_pred | 85 | 41 | BF397269 | similar to putative repair and recombination helicase RAD26L (predicted) |
| Smarca4 | 243 | 160 | BM385181 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
|
| ||||
| Transcriptional Regulation | ||||
|
| ||||
| Epc2_pre | 244 | 140 | AW918173 | enhancer of polycomb homolog 2 (predicted) |
| Pcgf2_pre | 99 | 655 | BI294621 | polycomb group ring finger 2 (predicted) |
| C. F3 Generation Regulated Epigenetic Associated Genes | ||||
| Microarray Signal | ||||
| Gene Category & Symbol | Control | Vinclozolin | Genbank # | Description/Function |
|
| ||||
| Histone Modification | ||||
|
| ||||
| RGD1566399_pre | 273 | 174 | BE098769 | Similar to MYST histone acetyltransferase monocytic leukemia 4 (predicted) |
| Hdac4_pre | 106 | 62 | BF419085 | Histone deacetylase 4 (predicted) |
| Ehmt1_pre | 508 | 330 | BM389055 | Euchromatic histone methyltransferase 1 (predicted) |
| LOC49817 | 215 | 143 | Al235220 | Similar to SET domain-containing protein |
| Hist1h4b | 86 | 144 | BM986536 | Germinal histone H4 gene |
|
| ||||
| Chromatin Remodeling | ||||
|
| ||||
| Smarca2 | 90 | 144 | BF547582 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| Chd7_pre | 100 | 66 | AI599104 | Chromodomain helicase DNA binding protein 7 (predicted) |
Statistically significant (P<0.05) changes (>1.5 fold) in gene expression with the bold genes having a decrease in the vinclozolin generation E16 testis and the italic gene is in common between the F1, F2 and F3 generations.
To confirm the microarray analysis a semi-quantitative PCR procedure was used to examine Dnmt1, Dnmt3A and Ehmt1 expression. Dnmt3L is a germ cell specific isoform of DNA methyltransferase [17; 18] that was not on the microarray, so was analyzed with the PCR procedure. The E16 testis from control and vinclozolin F1-F3 generation rats were collected and analyzed. The semi-quantitative PCR procedure was performed with three different experiments and three different RNA samples. The analysis confirmed the microarray observations and demonstrated the transient F1 generation decrease in the expression of Dnmt1, Dnmt3A Dnmt3L and Ehmt1, Figure 5. Therefore, the microarray data was confirmed with this semi-quantitative PCR analysis of Dnmt1, Dnmt3A and Ehmt1. The Dnmt3L germ cell specific isoform was decreased in a similar manner to the Dnmt3A in the F1 and F2 generation. Interestingly, some genes (e.g. Dmnt3A) were transiently decreased in the F1 or F2 generation and returned to normal levels by the F3 generation, while others (e.g. Dmnt3L) were decreased for all generations.
Figure 5.
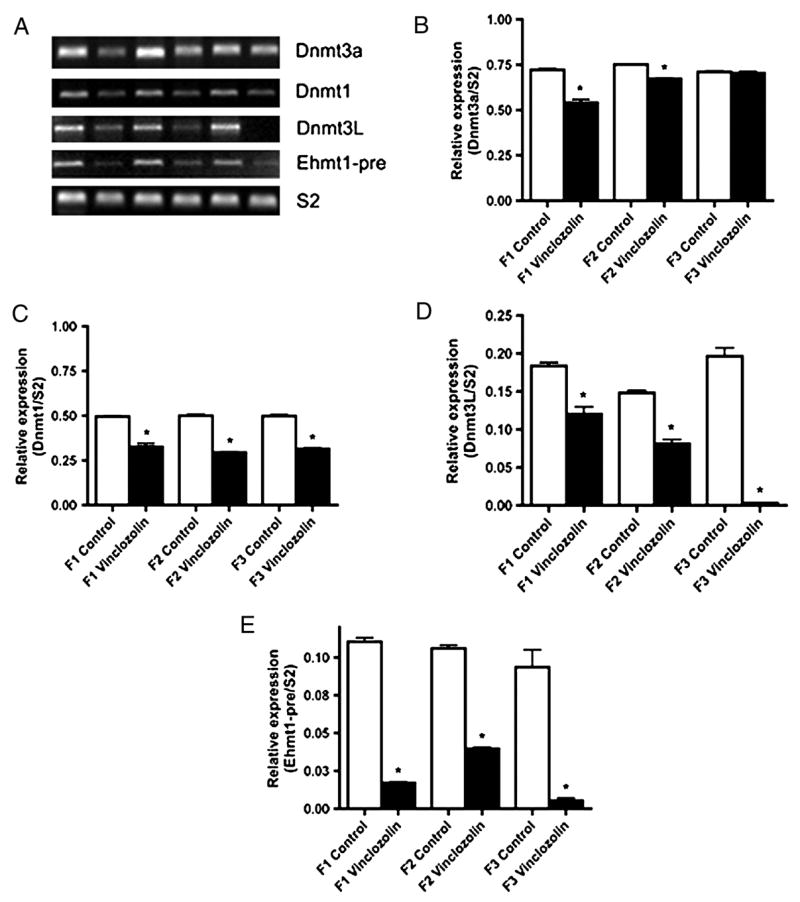
Semi-quantitative PCR analysis of (A & B) Dnmt3A, (A & C) Dnmt1, (A & D) Dnmt3L, (A & E) Ehmt1, and (A) the constitutively expressed S2. A representative electrophoretic gel of the PCR products is presented (A) and the combined data from 3 different experiments with normalization to S2 presented (B–E). The mean ± SEM is presented with asterisks (*) indicating a statistical difference (p<0.05) between control and vinclozolin generation F1-F3 E16 expression levels.
The hypothesis that “vinclozolin exposure during the transition from hypomethylation to hypermethylation of the fetal germ cells re-programs the epigenome of the male germ-line” is speculated to promote the transcriptome effects and adult onset disease phenotype to become transgenerational. Therefore, a select group of genes previously shown to be involved in epigenetic regulation of the genome were analyzed including genes associated with DNA methylation, histone modification and chromatin remodeling. Those genes involved in the epigenetic regulation and found to have an altered expression in the F1, F2 or F3 generation E16 testis are shown in Table 1. Interestingly, 19 epigenetic associated genes had altered expression in the F1 vinclozolin generation compared to control (Table 1A). The F2 vinclozolin generation E16 testis had 17 genes altered with 8 genes in common with F1 and 1 gene common with F3, Table 1B. Seven distinct epigenetic associated genes were altered in the F3 generation, (Table 1C). Genes were represented in each category and only one, Ehmt1, was in common between F1, F2 and F3 vinclozolin generations. Observations suggest the direct actions of vinclozolin on the F1 generation effects an increased number of epigenetic associated genes, which are speculated to be involved in the reprogramming of the germ-line epigenome. In contrast, the F3 generation altered genes are distinct and associated with the transgenerational tanscriptome and phenotype, but not necessarily involved in the direct actions of vinclozolin. Therefore, the actions of vinclozolin on the F1 generation appears to modify the epigenetic reprogramming to in part promote a permanent change (e.g. imprinting-like status) that causes epigenome alterations to be transmitted transgenerationally to subsequent F3 generations. The transcriptome changes in the F1 versus F3 generation contain some gene alterations in common, but many that are distinct.
DISCUSSION
Endocrine Disruptor Actions
Endocrine disruptors are a class of compounds that alter hormone actions and endocrinology [8; 9; 10]. Many environmental compounds from fungicides to plastics have endocrine disruptor activity [10; 20]. Embryonic and early postnatal exposure to these compounds has been shown to promote adult onset disease in numerous species [1; 6; 9; 20; 21]. The frequency of transgenerational disease induction and reproducibility of the phenomena [6; 7] suggests DNA sequence mutations are not likely the causal factor. A mechanism to consider is an alteration in the epigenetic programming (e.g. DNA methylation) of the genome. Epigenetics (e.g. DNA methylation) has been shown to have a significant impact on the regulation of gene expression and the transcriptome [22; 23]. The possibility that these environmental compounds promote an alteration in the epigenome (i.e. DNA methylation pattern) that then modifies the transcriptome associated with disease development was investigated in the current study.
Two potential epigenetic sites of action for environmental toxicants need to be considered. The first is during the active development of a specific organ when the epigenome and transcriptome is going through a cascade of developmental stages to eventually establish the adult organ transcriptome and physiology. In the event a factor reprogrammed or altered an epigenetically labile site or metastable allele of the epigenome during this development, the adult organ may not have the proper transcriptome and become susceptible to develop disease. This epigenetic induced adult onset disease state would not be passed to subsequent generations, but may be a significant factor in disease etiology [24]. The second major epigenetic site of action involves reprogramming the epigenome of the germ-line [6; 8]. The embryonic programming of the genome during sex determination could be modified to promote an abnormal epigenome. In the event this modified epigenetic program (i.e. DNA methylation) became imprinted then all subsequent generation programming would be influenced. Since the primordial germ cells undergo a de-methylation prior to sex determination and then re-methylation in a sex specific manner [13; 14; 15] during gonadal sex differentiation, the germ cells at this time could be sensitive to epigenetic re-programming [6; 7; 8]. This action of environmental factors could promote a transgenerational phenotype if the germ-line reprogramming was inherited [5; 8]. Observations demonstrate that the endocrine disruptor vinclozolin can alter the epigenome of the male germ-line transgenerationally. The ability of this altered epigenome to promote a transgenerational epigenetic alteration in the transcriptome is speculated to be the mechanism behind the subsequent development of heritable adult onset disease [7].
Transgenerational Transcriptome Regulation
As a model organ the embryonic testis was selected to examine the transgenerational epigenetic transcriptome. The F1, F2 and F3 control and vinclozolin generation E16 testis was obtained for analysis. Although morphological abnormalities were observed in adult animals [6], no histological changes were observed in the E16 testis. As seen with the adult animals [6], the vinclozolin F1 and F2 generation E16 testis also had increased germ cell apoptosis, but the F3 generation did not. Interestingly, the transcriptomes of F1-F3 vinclozolin generation E16 testis were significantly altered. A set of 196 genes were found to be consistently affected in the F1-F3 generation animals. Since the F1 generation embryo and F2 generation germ-line are directly exposed to vinclozolin, the number of genes affected is speculated to be higher in these generation E16 testis. The F3 generation is the first that is directly transgenerational (i.e. not having direct exposure) and the observations demonstrate the presence of a transgenerational epigenetic effect on the transcriptome. Developing organs have a cascade of changes in the epigenome and transcriptome that lead to the adult stage of differentiation. Therefore, the transgenerational epigenetic changes in the transcriptome will be distinct at different developmental stages, and distinct for different organs and cell types.
Sites of Potential Epigenetic Regulation
Although a number of different epigenetic regulatory mechanisms exist, the primary heritable epigenetic mechanism involves DNA methylation. The potential role of other epigenetic mechanisms (e.g. histone modification) remains to be thoroughly established. DNA methylation patterns are established during embryogenesis through the cooperation of methyltransferases and associated proteins [16]. Dnmt1 is responsible for maintenance of methylation patterns throughout DNA replication (i.e. specific to hemi-methylated sequences) and is localized in nuclear somatic tissues [25]. Dnmt2 may be involved in embryonic stem cells [26] and potential RNA methylation [27]. Dnmt3A and Dnmt3B are involved in active de novo DNA methylation at CpG sites [28]. Dnmt3 and Dnmt1 can cooperate and interact to regulate DNA methylation [29]. Dnmt3A interaction with Dnmt3L, the germ-line specific Dnmt, appears to be responsible for imprinting of the germ-line [29]. Dnmt3A has been shown to influence imprinting of a number of genes in the germ-line [28; 29; 30; 31]. Although Dnmt3L does not alone have methyltransferase activity, abnormal expression of Dnmt3L alters imprinting and allelic gene expression [18; 32]. A heterodimer of Dmnt3A and Dmnt3L appears to have a role in DNA methylation and the structure suggests unique specificity for the methylation of imprinted genes in the germ-line [33]. The current study demonstrates vinclozolin exposure during sex determination alters the expression of Dnmt’s in the F1 and F2 generations, with a return to more normal levels by the F3 generation. A dramatic effect on Dnmt3A and Dnmt3L was observed and correlates with its role in imprinting of the germ-line [19]. Therefore, alterations in the epigenome of the primordial germ cell and E16 testis transcriptome are speculated to involve in part alterations in Dnmt expression and activity. The transgenerational nature of the phenotype by the F3 generation likely does not directly involve Dnmt’s, but possible permanent alterations in the epigenome of the germ-line. This is related to the proposal that vinclozolin promotes an epigenetic reprogramming of the male germ-line involving an altered DNA methylation on a newly induced imprinted-like DNA sequences [5; 6]. In the F1 generation exposure the germ-line is reprogrammed to develop these new imprinted-like sites that then transgenerationally transmit the alteration in the male germ-line epigeneome to subsequent generations. A known imprinted gene (i.e. H19) was found not to have a change in methylation after vinclzolin exposure, such that the reprogramming may involve the induction of new sites [34]. Therefore, the epigenetic transgenerational phenotype initially involves effects on the DNA methylation machinery (i.e. methyltransferases) and then appears transgenerationally transmitted through permanent changes in the epigenome.
A combination of factors and proteins are likely involved in the altered epigenetic programming observed, and not simply reflected in a regulation of DNA methyltransferases alone. Additional genes associated with epigenetic regulation were shown to be altered in the F1 vinclozolin generation E16 testis. In contrast, a small distinct set of epigenetic associated genes were altered in the F3 vinclozolin generation, with only one gene (i.e. Ehmt1) in common between the F1, F2 and F3 generations. All three major epigenetic categories of DNA methylation, histone modification and chromatin remodeling were represented in the effected gene list. An interesting example of a histone methyltransferase analyzed was euchromatic histone methyltransferase 1 (Ehmt1), which can promote histone methylation to suppress transcription [35] and indirectly influence DNA methylation [36]. Ehmt1 expression in the F1-F3 vinclozolin generation E16 testis was found to decrease which suggests a combination of DNA and histone methyltransferases will likely be involved in the initial actions of vinclozolin. Therefore, the direct exposure to vinclozolin in the F1 generation altered the expression of a significant number of epigenetic associated genes that are presumed to in part be responsible for the re-programming of the germ-line epigenome. Those epigenetic associated genes altered in the F3 vinclozolin generation are likely associated with the transgenerational transcriptome. Observations suggest the methylation and epigenetic machinery is modified after vinclozolin exposure, but the specific mechanisms involved in the reprogramming of the germ-line epigenome remain to be elucidated.
Previously, we have shown that the endocrine disruptor vinclozolin after embryonic exposure prior to and during gonadal sex differentiation promotes the adult onset disease involving spermatogenic cell defects and male fertility [6], as well as a variety of other disease states [6]. Investigation of the transgenerational sperm epigenome identified genes/DNA sequences with potential methylation characteristics in F3 vinclozolin generation animals [7]. The current study demonstrates a transgenerational effect on the transcriptome of the developing embryonic testis. Therefore, the mechanism proposed is that the environmental factor (i.e. endocrine disruptor vinclozolin) acts on the embryonic gonad during gonadal sex determination/differentiation to alter the epigenetic programming of the male germ-line [6; 7; 8]. This altered germ-line epigenome appears to in a transgenerational manner subsequently modify the transcriptomes of developing organs and is speculated to result in adult onset disease. The epigenetic effects of environmental exposures could provide a mechanism for many toxicant exposure phenotypes observed [5]. Although the level of vinclozolin used in the current study exceeds that observed in the environment, such that no conclusion on toxicology can be made, the phenomenon of developing a transgenerational epigenetic effect on the transcriptome is critical to understand the potential mechanisms involved in toxicology and disease. The ability of an environmental factor to influence the transgenerational epigenetic programming of an organs transcriptome provides a critical factor in disease etiology not previously considered. Further analysis of this transgenerational epigenetic transcriptome now requires individual organ systems and disease states to be investigated.
MATERIALS & METHODS
A. In Vivo Procedures
Gestating outbred Sprague-Dawley mother rats from timed pregnant colonies housed at the Washington State University Vivarium were given intraperitoneal (IP) injections of vinclozolin (100mg/kg/day) from embryonic day 8–14 (E8-E14) of gestation (i.e. F0 generation) as previously described [37]. Sperm positive vaginal smear date being embryonic day 0. Gestating control mothers (i.e. F0 generation) received vehicle alone (i.e. sesame oil and DMSO). At least 8 lines (individual F0 injected females) were generated for controls and 8 lines for vinclozolin generations for these analyses. The F1-F3 generation animals derived from a vinclozolin exposed F0 mother are referred to as vinclozolin generation animals, while those from control F0 mothers are identified as control generation animals. The testes from male rats from control and vinclozolin generations were collected at E16 for analysis. Adult F1 vinclozolin generation (offspring from F0 mothers) males were bred to F1 vinclozolin generation females to generate the F2 vinclozolin generation. Adult F2 vinclozolin generation males were bred to F2 vinclozolin generation females to generate the F3 vinclozolin generation. Rats for the control groups (i.e. generations F1-F3) were bred in the same manner for all the generations. No inbreeding or sibling crosses were generated. The vinclozolin outcross group (VOC) was generated by breeding the vinclozolin F2 generation males with wild-type females (total of 6 litters). Wildtype E16 litters were collected from wildtype male and female breedings. Pregnant wildtype females did not receive any injections and were used to verify the effects of the treatment. All procedures have been approved by the Washington State University Animal Use and Care Committee.
B. Histology
Tissues were fixed in Bouin’s (Sigma St. Louis, MO) for 1 hour, embedded in paraffin and sectioned. Sections were stained with hematoxylin and eosin according to standard procedures. The Center for Reproductive Biology, Histology Core Laboratory assisted with these procedures. The animal numbers were n= 6 for vinclozolin and n=6 for controls from 3 different lines (i.e. F0 injected mothers) from each generation (F1-F3).
C. Detection of Cell Apoptosis
To detect apoptotic cells in testis sections, the Fluorescein In Situ Cell Death Detection Kit (Roche Applied Science, Indianapolis, IN) was utilized [37]. This system measures the fragmented DNA from apoptotic cells by enzymatically incorporating fluorecein-12-dUTP at the 3′-OH DNA ends using the enzyme terminal deoxynucleotidyl transferase which forms a polymeric tail using the principle of the TUNEL assay. Fluorescent apoptotic cells were imaged on a confocal microscope and number of apoptotic cells per testis cross-section determined. A minimum of n=6 for vinclozolin and n=4 for controls for each generation was used. All cross sections used for TUNEL analysis had normal testis morphology.
D. Microarray Analysis and Bioinformatics
RNA was hybridized to the Affymetrix (Affymetrix, Santa Clara, California) rat 230 2.0 gene chip. The Genomics Core in the Center for Reproductive Biology at Washington State University performed the analysis as previously described [38; 39]. Briefly, RNA from control and vinclozolin generation E16 testis was reverse transcribed into cDNA and cDNA was transcribed into biotin labeled RNA. Biotin labeled RNA was then hybridized to the Affymetrix rat 230 2.0 gene chips. Biotinylated RNA was then visualized by labeling with phycoerythrin-coupled avidin. The microarray chip was scanned on a Affymetrix Gene Chip Scanner 3000 (Affymetrix, Santa Clara, CA). The microarray image data were converted to numerical data with GeneChip Operating Software (GCOS version 1.1; Affymetrix) using a probe set scaling factor of 125. An absolute analysis was performed with GCOS to assess the relative abundance of the transcripts based on signal and detection calls (present, absent or marginal). This information was imported into Genespring software (Silicon Genetics, Redwood City, CA) and normalized using the recommended defaults. This includes setting signal values below 0.01 to a value of 0.01, total chip normalization to the 50th percentile, and normalization of each chip to the median. Unless otherwise indicated, in order for a transcript to be considered present it had to be both flagged as present in the GCOS present/absent call, and have an expression level greater than 75. Briefly, the 16 sets of oligonucleotides for a specific gene were used to make comparisons of a signal to statistically determine a present call using a one-sided Wilcoxon’s signed rank test. In order for a transcript to be considered changed between treatment groups it had to exhibit at least a 1.5-fold change between the means of the treatments and have a Students t-test p-value p≤0.05 between treatments. The raw signal cut off was between 75. Therefore, the data presented are for genes that were determined to be statistically present and found to have a statistically different change.
Two different experiments were performed involving two different sets of animals, RNA sample preparations and microarray chips. Therefore, two control and vinclozolin generation E16 testis samples were analyzed on two different chips. This allowed a 2×2 factorial comparison with all present/absent calls and changes in expression to be statistically significant for further analysis. The R2 for the comparison between microarray chips was found to be R2 > 0.94, which indicated negligible total variability between chips, experiments and samples. This R2 value and statistical analysis indicated the chip number used was appropriate. The number of chips required for specific experiments has been previously reviewed [40]. Previous studies have demonstrated that microarray data is validated with quantative PCR data [39; 41]. Due to the presence of 16 different oligonucleotide sets for each specific gene being used on the microarray versus only a single primer set for a gene in a quantative PCR, the microarray is more effective at eliminating false positive or negative data and provides a more robust quantitation of changes in gene expression. However, validation of microarray data was performed with selected genes using a semi-quanitative PCR procedure. Although the magnitude of the change can vary, the absence or presence of a change is generally consistent. A number of the methyltransferases (i.e. Dnmt1, Dnmt3L, Dnmt3A and Ehmt1) were selected to perform a semi-quantitative PCR analysis to confirm the microarray data. The primer sets used for the genes were (Dnmt1, forward 5′-GTGGGATGGCTTCTTCAGTA-3′ and reverse 5′-GGCTTGGTCACAAAACAAAC-3′), (Dnmt3L, forward 5′-CGCTGAAGTACGTGGAAGAT-3′ and reverse 5′-ACTTGGGTTTGCAGAGACTG-3′), (Dnmt3A, forward 5′-TTGGCTTCCCTGTCCACTAC-3′ and reverse 5′-ATGATGTCCAACCCTTCTGC-3′), (Ehmt1, forward ‘5-ATGTAAATGGCGAGAGCTTG-3’, and reverse 5′-TTCCTGGGGATGACTTACAA-3′ and (S2, forward ‘5- GCTCGTGGAGGTAAAGCTGA-3’, and reverse 5′-TGAGACGAACCAGCACAGAG-3′). Similar observations were made with this semi-quantitative PCR procedure and the microarray analysis.
F. Statistical Analysis
The data from apoptotic cell numbers were analyzed using a SAS program. The values were expressed as the mean ± SEM. Statistical analysis was performed and the difference between the means of treatments and respective controls was determined using a Student’s t-test. In vivo experiments were repeated with 3–6 individuals for each data point. A statistically significant difference was confirmed at p < 0.05.
Supplementary Material
Acknowledgments
We acknowledge the technical contributions of Dr. Ingrid Sadler-Riggleman, Mr. Trevor Covert, and the assistance of Dr. Marina Savenkova in the Washington State University, Center for Reproductive Biology Bioinformatics Core Laboratory, and Mr. Derek Pouchnik in the Genomics Core Laboratory for assistance with the bioinformatics. This research was supported by a grant from the National Institute of Environmental Health Sciences, NIH to MKS.
Footnotes
Note: All three authors contributed equally to the manuscript.
Disclosure: None of the authors have a financial or conflict of interest to disclose
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gluckman PD, Hanson MA. Developmental origins of disease paradigm: a mechanistic and evolutionary perspective. Pediatr Res. 2004;56:311–7. doi: 10.1203/01.PDR.0000135998.08025.FB. [DOI] [PubMed] [Google Scholar]
- 2.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–73. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 3.Jiang YH, Bressler J, Beaudet AL. Epigenetics and human disease. Annu Rev Genomics Hum Genet. 2004;5:479–510. doi: 10.1146/annurev.genom.5.061903.180014. [DOI] [PubMed] [Google Scholar]
- 4.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 5.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–62. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–9. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anway MD, Leathers C, Skinner MK. Endocrine Disruptor Vinclozolin Induced Epigenetic Transgenerational Adult Onset Disease. Endocrinology. 2006;147:5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anway MD, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors. Endocrinology. 2006;147:S43–9. doi: 10.1210/en.2005-1058. [DOI] [PubMed] [Google Scholar]
- 9.Crews D, McLachlan JA. Epigenetics, evolution, endocrine disruption, health, and disease. Endocrinology. 2006;147:S4–10. doi: 10.1210/en.2005-1122. [DOI] [PubMed] [Google Scholar]
- 10.Fisher JS. Environmental anti-androgens and male reproductive health: focus on phthalates and testicular dysgenesis syndrome. Reproduction. 2004;127:305–15. doi: 10.1530/rep.1.00025. [DOI] [PubMed] [Google Scholar]
- 11.Kelce WR, Monosson E, Gamcsik MP, Laws SC, Gray LE., Jr Environmental hormone disruptors: evidence that vinclozolin developmental toxicity is mediated by antiandrogenic metabolites. Toxicol Appl Pharmacol. 1994;126:276–85. doi: 10.1006/taap.1994.1117. [DOI] [PubMed] [Google Scholar]
- 12.Uzumcu M, Suzuki H, Skinner MK. Effect of the anti-androgenic endocrine disruptor vinclozolin on embryonic testis cord formation and postnatal testis development and function. Reprod Toxicol. 2004;18:765–74. doi: 10.1016/j.reprotox.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 14.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet 14 Spec No. 2005;1:R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 15.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 16.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 17.Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–9. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 18.Webster KE, O’Bryan MK, Fletcher S, Crewther PE, Aapola U, Craig J, Harrison DK, Aung H, Phutikanit N, Lyle R, Meachem SJ, Antonarakis SE, de Kretser DM, Hedger MP, Peterson P, Carroll BJ, Scott HS. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc Natl Acad Sci U S A. 2005;102:4068–73. doi: 10.1073/pnas.0500702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–3. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 20.Heindel JJ. The fetal basis of adult disease: Role of environmental exposures--introduction. Birth Defects Res A Clin Mol Teratol. 2005;73:131–2. doi: 10.1002/bdra.20119. [DOI] [PubMed] [Google Scholar]
- 21.Newbold RR, Hanson RB, Jefferson WN, Bullock BC, Haseman J, McLachlan JA. Increased tumors but uncompromised fertility in the female descendants of mice exposed developmentally to diethylstilbestrol. Carcinogenesis. 1998;19:1655–63. doi: 10.1093/carcin/19.9.1655. [DOI] [PubMed] [Google Scholar]
- 22.Barber R, Plumb MA, Boulton E, Roux I, Dubrova YE. Elevated mutation rates in the germ line of first- and second-generation offspring of irradiated male mice. Proc Natl Acad Sci U S A. 2002;99:6877–82. doi: 10.1073/pnas.102015399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong H, Bonala RR, Suzuki N, Johnson F, Grollman AP, Shibutani S. Mutagenic potential of benzo[a]pyrene-derived DNA adducts positioned in codon 273 of the human P53 gene. Biochemistry. 2004;43:15922–8. doi: 10.1021/bi0482194. [DOI] [PubMed] [Google Scholar]
- 24.Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–32. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leohnardt H, Bestor TH. Structure, function and regulation of mammalian DNA methyltransferase. In: Jost JPSHP, editor. DNA Methylation: Molecular Biology and Biological Significance. Berkauser; Berlin: 1993. pp. 109–119. [DOI] [PubMed] [Google Scholar]
- 26.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22:480–91. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–8. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 28.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–20. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 29.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci U S A. 2002;99:16916–21. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 31.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–23. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 32.Bourc’his D, Le Bourhis D, Patin D, Niveleau A, Comizzoli P, Renard JP, Viegas-Pequignot E. Delayed and incomplete reprogramming of chromosome methylation patterns in bovine cloned embryos. Curr Biol. 2001;11:1542–6. doi: 10.1016/s0960-9822(01)00480-8. [DOI] [PubMed] [Google Scholar]
- 33.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007 doi: 10.1038/nature06146. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang HS, Anway MD, Rekow SS, Skinner MK. Transgenerational epigenetic imprinting of the male germ-line by endocrine disruptor exposure during gonadal sex determination. Endocrinology. 2006;147 doi: 10.1210/en.2006-0987. [DOI] [PubMed] [Google Scholar]
- 35.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–91. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikegami K, Iwatani M, Suzuki M, Tachibana M, Shinkai Y, Tanaka S, Greally JM, Yagi S, Hattori N, Shiota K. Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells. Genes Cells. 2007;12:1–11. doi: 10.1111/j.1365-2443.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- 37.Cupp AS, Uzumcu M, Suzuki H, Dirks K, Phillips B, Skinner MK. Effect of transient embryonic in vivo exposure to the endocrine disruptor methoxychlor on embryonic and postnatal testis development. J Androl. 2003;24:736–45. doi: 10.1002/j.1939-4640.2003.tb02736.x. [DOI] [PubMed] [Google Scholar]
- 38.McLean DJ, Friel PJ, Pouchnik D, Griswold MD. Oligonucleotide microarray analysis of gene expression in follicle-stimulating hormone-treated rat Sertoli cells. Mol Endocrinol. 2002;16:2780–92. doi: 10.1210/me.2002-0059. [DOI] [PubMed] [Google Scholar]
- 39.Shima JE, McLean DJ, McCarrey JR, Griswold MD. The Murine Testicular Transcriptome: Characterizing Gene Expression in the Testis During the Progression of Spermatogenesis. Biol Reprod. 2004 doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 40.Chen JJ, Delongchamp RR, Tsai CA, Hsueh HM, Sistare F, Thompson KL, Desai VG, Fuscoe JC. Analysis of variance components in gene expression data. Bioinformatics. 2004;20:1436–46. doi: 10.1093/bioinformatics/bth118. [DOI] [PubMed] [Google Scholar]
- 41.Kezele PR, Ague JM, Nilsson E, Skinner MK. Alterations in the ovarian transcriptome during primordial follicle assembly and development. Biol Reprod. 2005;72:241–55. doi: 10.1095/biolreprod.104.032060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






