Abstract
We present a simple and inexpensive ‘one-step’ protocol for the hydrolysis of DNA to deoxyribonucleosides. Unlike the older DNA hydrolysis protocol which is cumbersome and labor intensive, thes new protocol is ideal for high-throughput assays and is suitable automation. Using this protocol we were able to hydrolyze several hundred samples within an 8-hour period. The new protocol is fully compatible with LC-MS/MS and gives similar recoveries for all five major deoxyribonucleosides when compared to the older protocol.
The requirement to hydrolyze DNA to deoxyribonucleosides is a common component of several assays. To measure the turnover of DNA [1, 2] or cells [3–5] in various tissues, DNA can be labeled with isotope-labeled precursor. In such studies, the DNA is extracted, hydrolyzed, and analyzed to determine label incorporation by mass spectrometry. Likewise, in vivo experiments in humans, such as examination of the effect of gene polymorphisms [6] and nutritional status [6, 7] on DNA metabolism, have used labeled one-carbon donors to label monocyte DNA. Nucleoside hydrolysates of DNA also have been used in determining the base composition of DNA, including investigation of epigenetic modification such as deoxycytosine methylation [8, 9], oxidative damage [10, 11], or unique DNA composition [12].
One of the most commonly used protocols for digesting DNA is the tri-enzyme protocol devised by Crain [13]. However, a major weakness of the Crain protocol is that the first enzyme in the reaction, nuclease P1 (E.C. 3.1.30.1), only can digest single stranded DNA and has a pH optimum (pH ~5) significantly lower and a reaction temperature significantly higher (~50°C) than the other two enzymes (pH >8 and ~37°C) in the reaction. This leads to an overly complex protocol [Fig 1], whereby the DNA must be boiled to denature (and rapidly cooled to prevent reversion), the pH of the sample is adjusted twice, and the sample undergoes three separate enzyme incubations. These complexities limit the number of samples that can be prepared. For logistical reasons (repetitive manipulations of tubes), we were typically [6] restricted to preparing ~60 samples per day when we used the Crain protocol. Furthermore, the Crain protocol uses high buffer concentrations (e.g., 50–100 mmol/L ammonium bicarbonate, pH 7.9) which can interfere with downstream reactions and assays. Therefore, we devised a simple and inexpensive protocol for hydrolyzing DNA to deoxynucleosides that can be performed in a single incubation, without the need to denature the DNA.
Fig 1.
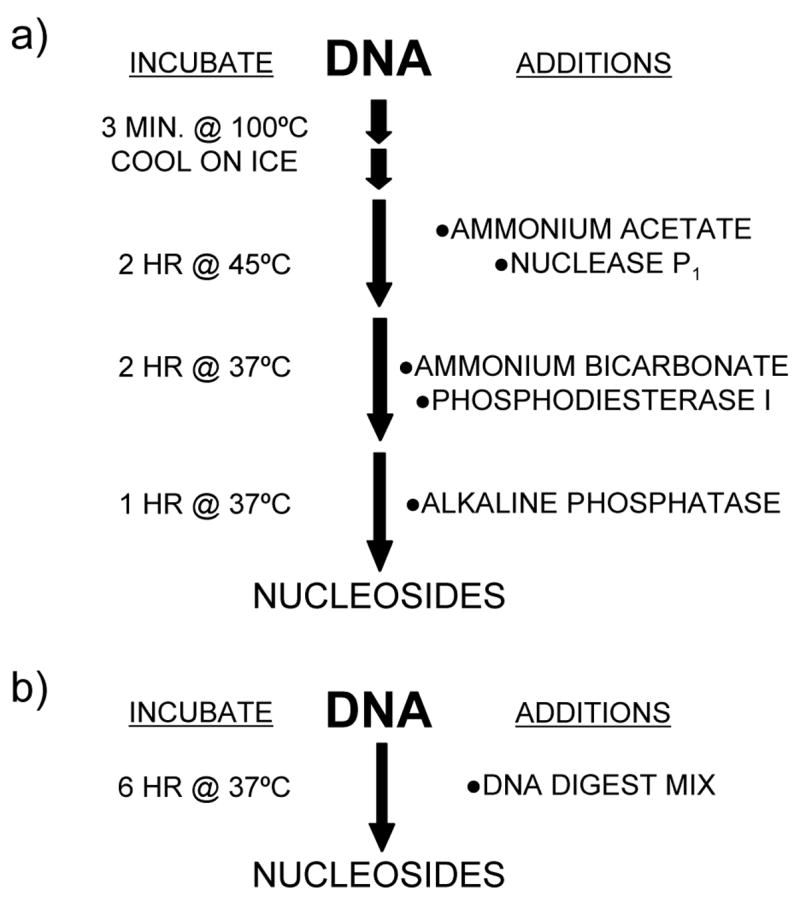
DNA digestion flow diagram showing the method of a) Crain [13], and b) our revised method.
This was achieved by substituting an endonuclease from Serratia marcescens (E.C. 3.1.30.2) for nuclease P1. This enzyme exhibits no preference for single or double stranded DNA, and has pH (~pH 8) and temperature (~37°C) optima similar to the other enzymes in the reaction. We use a commercial variant of the endonuclease enzyme, Benzonase (Merck, Darmstadt, Germany), produced by over-expressing the recombinant enzyme in E. coli. Benzonase is sold commercially as a reagent for purifying protein by removing DNA and RNA.
A typical DNA digestion protocol is described: DNA is extracted and purified using standard laboratory protocols or a commercial kit. The DNA is reconstituted in water or DNA hydrolysis buffer at a typical concentration of 20 ng/μL. Digest Mix (enough for one hundred 1μg samples) was prepared by adding 250 Units Benzonase, 300 mUnits phosphdiesterase I (Sigma P-3243) and 200 Units alkaline phosphatase (Sigma P-7923) to 5 mL Tris-HCl buffer (20mM, pH7.9) containing 100 mM NaCl and 20 mM MgCl2. DNA samples (1 μg) were digested by adding 50 μL Digest Mix. After incubating at 37°C for 6 hr the DNA was ready for analysis [Fig. 1b]. To determine completeness of hydrolysis, triplicate 1 μg samples of human placental DNA (Sigma D-7011) were digested for 6 hr, 8 hr, or overnight (~18 hr). Samples were chromatographed on a Luna C18(2) column (50 × 3.0 mm, 3μ; Phenomenex, Torrence CA) eluted at 0.3 mL/min with 1 mmol/L ammonium acetate/0.1% acetic acid using a methanol (4% to 12%) gradient. Tandem mass spectrometry was conducted using a Termo Finnigan TSQ Quantum mass spectrometer in H-ESI mode (200°C heated vaporizer) using the mass transitions previously reported [6]. Incubation time had no effect on peak area. No attempt was made to further optimize incubation times or enzyme concentrations.
For comparison 1μg human placental DNA (in 50μl water) was digested using the Crain protocol [13]. In brief, the DNA sample was denatured by boiling in a water bath at 100°C for 3 min. The sample was cooled rapidly on ice before 5 μL ammonium acetate (0.1 mol/L, pH 5.3) was adding. After incubating with 2 U nuclease P1 (Sigma N-8630) at 45°C for 2 hr the pH of the sample was adjusted with 5 μl fresh ammonium bicarbonate (1 mol/L) before 3 mUnits phosphodiesterase I were added and incubated at 37°C for a further 2 hr. After incubating with 2 Units alkaline phosphatase at 37°C for 1 hr the volume of the sample was made up to 100 μL with water. Components of the Crain digestion (presumably the high ammonium bicarbonate and/or ammonium acetate concentration) interfered with both the sample chromatograph and ionization (increasing diamerization of deoxycytidine, and of deoxyadenosine). Consequently, the composition of both samples was adjusted to give a similar buffer composition. Specifically, the sample digested using the Crain protocol was diluted with 100 μL Tris-HCl buffer (10mM, pH7.9) containing 50 mM NaCl and 10 mM MgCl2; the sample digested using our new protocol was diluted with 50 μL ammonium acetate (10 mmol/L, pH 5.3) and 50 μL ammonium bicarbonate (100 mmol/L). Both samples were then diluted with 1.8 ml water (spiked with [15N3]deoxycytidine (Cambridge Isotopes, Cambridge MA) and [2H4]thymidine (Cambridge Isotopes) as internal standards). Both digestion protocols gave comparable deoxyribonucleoside concentrations [Table 1], particularly when the deoxycytidine peak area was corrected using recovery of its internal standard.
Table 1.
Comparison of digestion methods. Human placental DNA was digested using either our new protocol or the Crain protocol [13], see text for methods. Each sample was injected 6 times.
| dCyt 1 | [15N]dCyt | dCyt ratio2 | MdCyt | dThy | [2H]dThy | dThy Ratio3 | dAdn | dGua | ||
|---|---|---|---|---|---|---|---|---|---|---|
| New | Mean | 402,195 | 261,561 | 1.54 | 62,198 | 589,904 | 474,169 | 1.24 | 6,556,581 | 780,122 |
| %RSD | 11.1 | 12.6 | 4.5 | 7.0 | 4.5 | 4.9 | 2.1 | 5.0 | 4.6 | |
| Crain | Mean | 333,190 | 231,026 | 1.44 | 61,095 | 594,360 | 484,428 | 1.16 | 6,454,096 | 797,842 |
| %RSD | 5.7 | 6.1 | 4.7 | 5.2 | 4.9 | 4.0 | 2.0 | 9.2 | 5.6 | |
|
| ||||||||||
| %Difference4 | −17.2 | −11.7 | −6.3 | −1.8 | −4.3 | 2.2 | −6.4 | −1.6 | 2.3 | |
dCyt: deoxycytidine; MdCyt: 5-methyldeoxycytidine; dThy: thymidine; dAdn; deoxyadenosine; dGua: deoxyguanosine.
Ratio of deoxycytidine to its [15N3]labeled internal standard.
Ratio of thymidine to its [2H4]labeled internal standard.
Percentage difference between the mean values derived using the new protocol or the Crain protocol.
In conclusion, we present a simple, one-step protocol for the digestion of DNA to deoxynucleosides that is suitable for automation. Using a repeating pipette, it has taken us less than two hours to prepare 450 samples for incubation. Reagent concentrations and digestion volumes are scalable, depending on the sample DNA concentration and the desired final volume. As all the enzymes used in the protocol are active towards RNA the protocol also could be used to hydrolyze RNA to ribonucleosides.
Acknowledgments
This research was supported in part by NIH grants DK37481 and DK56274, and funds from the Florida Agricultural Experiment Station.
Footnotes
Abbreviations Used: LC-MS/MS: liquid chromatography - tandem mass spectrometry; H-ESI: heated electrospray ionization; dCyt: deoxycytidine; MdCyt: 5-methyldeoxycytidine; dThy: thymidine; dAdn: deoxyadenosine; dGua: deoxyguanosine
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Busch R, Cesar D, Higuera-Alhino D, Gee T, Hellerstein MK, McCune JM. Isolation of peripheral blood CD4(+) T cells using RosetteSep and MACS for studies of DNA turnover by deuterium labeling. J Immunol Methods. 2004;286:97–109. doi: 10.1016/j.jim.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Collins ML, Eng S, Hoh R, Hellerstein MK. Measurement of mitochondrial DNA synthesis in vivo using a stable isotope-mass spectrometric technique. J Appl Physiol. 2003;94:2203–2211. doi: 10.1152/japplphysiol.00691.2002. [DOI] [PubMed] [Google Scholar]
- 3.Macallan DC, Fullerton CA, Neese RA, Haddock K, Park SS, Hellerstein MK. Measurement of cell proliferation by labeling of DNA with stable isotope-labeled glucose: Studies in vitro, in animals, and in humans. Proc Natl Acad Sci USA. 1998;95:708–713. doi: 10.1073/pnas.95.2.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neese RA, Misell LM, Turner S, Chu A, Kim J, Cesar D, Hoh R, Antelo F, Strawford A, McCune JM, Christiansen M, Hellerstein MK. Measurement in vivo of proliferation rates of slow turnover cells by 2H2O labeling of the deoxyribose moiety of DNA. Proc Natl Acad Sci USA. 2002;99:15345–15350. doi: 10.1073/pnas.232551499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz GN, Vance BA, Levine BM, Fukazawa M, Telford WG, Cesar D, Hellerstein M, Gress RE. Proliferation kinetics of subpopulations of human marrow cells determined by quantifying in vivo incorporation of [2H2]-glucose into DNA of S-phase cells. Blood. 2003;102:2068–2073. doi: 10.1182/blood-2003-01-0139. [DOI] [PubMed] [Google Scholar]
- 6.Quinlivan EP, Davis SR, Shelnutt KP, Henderson GN, Ghandour H, Shane B, Selhub J, Bailey LB, Stacpoole PW, Gregory JF. Methylenetetrahydrofolate reductase 677C -> T polymorphism and folate status affect one-carbon incorporation into human DNA deoxynucleosides. J Nutr. 2005;135:389–396. doi: 10.1093/jn/135.3.389. [DOI] [PubMed] [Google Scholar]
- 7.Quinlivan EP, Davis SR, Shelnutt KP, Henderson GN, Stacpoole PW, Gregory JF. The effects of marginal vitamin B6 deficiency on one-carbon incorporation into DNA deoxynucleosides. FASEB J. 2005;19:A54. doi: 10.1093/jn/135.3.389. [DOI] [PubMed] [Google Scholar]
- 8.Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nature Genetics. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 9.Friso S, Choi S-W, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R, Selhub J. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci USA. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ringvoll J, Nordstrand LM, Vågbø CB, Talstad Vivi, Reite K, Aas PA, Lauritzen KH, Liabakk NB, Bjørk A, Doughty RW, Falnes PØ, Krokan HE, Klungland Arne. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–2198. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishina Y, Yang CG, He C. Direct repair of the exocyclic DNA adduct 1,N-6-ethenoadenine by the DNA repair AlkB proteins. J Am Chem Soc. 2005;127:14594 –14595. doi: 10.1021/ja055957m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiljunen S, Hakala K, Pinta E, Huttunen S, Pluta P, Gador A, Lönnberg H, Skurnik M. Yersiniophage ΦR1-37 is a tailed bacteriophage having a 270 kb DNA genome with thymidine replaced by deoxyuridine. Microbiology. 2005;151:4093–4102. doi: 10.1099/mic.0.28265-0. [DOI] [PubMed] [Google Scholar]
- 13.Crain PF. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]


