Abstract
Deficiency in genes involved in DNA mismatch repair increases susceptibility to cancer, particularly of the colorectal epithelium. Using Msh2 null mice, we demonstrate that this genetic defect renders normal intestinal epithelial cells susceptible to mutation in vivo at the Dlb-1 locus. Compared with wild-type mice, Msh2-deficient animals had higher basal levels of mutation and were more sensitive to the mutagenic effects of temozolomide. Experiments using Msh2-deficient cells in vitro suggest that an element of this effect is attributable to increased clonogenicity. Indeed, we show that Msh2 plays a role in the in vivo initiation of apoptosis after treatment with temozolomide, N-methyl-N′-nitro-N-nitrosoguanidine, and cisplatin. This was not influenced by the in vivo depletion of O6-alkylguanine-DNA-alkyltransferase after administration of O6-benzylguanine . By analyzing mice mutant for both Msh2 and p53, we found that the Msh2-dependent apoptotic response was primarily mediated through a p53-dependent pathway. Msh2 also was required to signal delayed p53-independent death. Taken together, these studies characterize an in vivo Msh2-dependent apoptotic response to methylating agents and raise the possibility that Msh2 deficiency may predispose to malignancy not only through failed repair of mismatch DNA lesions but also through the failure to engage apoptosis.
Structural distortions produced by nucleotides that are either unpaired or paired with noncomplementary nucleotides are recognized by proteins encoded by the mismatch repair genes. Several members of this family of genes have been characterized within Saccharomyces cerevisiae, which led to the identification of mammalian homologues. Six human mismatch repair genes have been cloned: MSH2, MLH1, PMS1, PMS2, MSH3, and GTBP (1, 2). Constitutive inactivation of these genes has been associated with the development of cancer, the best characterized relationship being between the inherited cancer-susceptibility syndrome of hereditary nonpolyposis colorectal cancer and germline mutations in MSH2, MLH1, and PMS2 (2, 3). Mice have been produced bearing targeted inactivations of the Mlh1, Msh2, Pms1, and Pms2 genes (4, 5). Homozygous mice from all of these mutant strains are viable but prone to the development of different types of neoplasia, predominantly of the lymphoid and intestinal lineages.
MSH2 recognizes mismatched nucleotides as a heterodimer with GTBP (6). In what is assumed to be a direct consequence of failure of this process, microsatellite instability is increased in murine Msh2 −/− cells (4). These data support the concept that failed recognition and repair of mismatch lesions leads directly to an increase in mutation frequency and thereby to malignancy. However, the consequences of MSH2 deficiency are complex, with Msh2 −/− cells also showing hyper-recombination and increased survival after exposure to methylating agents in vitro (4). This last property suggests that MSH2 status may affect the ability to undergo apoptosis, with increased resistance to methylating agents arising from failure to detect damage and hence initiate apoptosis. Results from several different tumor cell lines deficient in mismatch repair are consistent with this hypothesis (7). We have used embryonic stem (ES) cells and mice bearing a gene-targeted inactivation of Msh2 (4) to investigate the in vivo role of this protein in the prevention of mutation and in the initiation of apoptosis after DNA damage. We find that, in the murine small intestine, Msh2 deficiency reduces the apoptotic response to temozolomide, N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), and cisplatin. We also show that this response is normally p53-dependent. In the absence of Msh2, we find a dose-dependent increase in the in vivo mutation frequency, and we have performed in vitro experiments that suggest that a component of this increase may arise from the failure to engage death pathways.
METHODS
Mice.
All of the mice used in these studies were derived from a single outbred colony segregating for Ola/129, BALB/c, and SWR genomes. The percentage contribution of each genome type was not determined for each experimental cohort, but, wherever possible, littermate controls were used to minimize any background-dependent effect. The apoptosis studies described here have been minimally reproduced on an inbred C57BL/6 background, and similar results have been obtained (data not shown). The Dlb-1 assay was performed as described (8). The Dlb-1 assay is a specific locus assay in which somatic mutations are detected that inactivate the polymorphic genetic locus, Dlb-1, which encodes a lectin binding site. The assay is autosomal with the locus located on mouse chromosome 11. Two alleles of Dlb-1 have been characterized. The Dlb-1b allele specifies binding of the Dolichos biflorus agglutinin to intestinal epithelium. The Dlb-1a allele specifies binding to the vascular endothelium. In mice that are Dlb-1b/Dlb-1a, any inactivating mutation of the single Dlb-1b allele leads to loss of the ability to bind the lectin. After cell proliferation and clonal expansion, mutations that occurred in the stem cell population of the intestine result in the formation of a clone that does not bind a peroxidase conjugate of Dolichos biflorus agglutinin. These unstained clones are easily identified and quantified in stained wholemounts of the small intestine. To perform the Dlb-1 assay, experimental cohorts were derived by backcrossing Msh2-deficient animals derived from the breeding colony described above to two different C57BL/6 strains, one of which was homozygous for the Dlb-1a allele and one of which was homozygous for the Dlb-1b allele. Mice subsequently were intercrossed from these two lines to generate mice heterozygous at the Dlb-1 locus and segregating for all possible Msh2 genotypes.
Genotyping Mice by PCR.
Tail biopsies were boiled in 500 μl of lysis buffer (0.1M Tris, pH 8.5/0.005M EDTA/0.2% SDS/0.2M NaCl) for 15 min. A sample (1 μl) was used as template in a 50-μl three primer PCR reaction containing 5 μl of PCR buffer, 2.5 μl of 1% detergent, 2.5 μl each of primer (10 pmol/μl), 1 μl of dNTP (40 mM), 2 μl of MgCl2 (50 mM), 30 μl of double-distilled water, and 1 μl of Taq (1.25 units) (GIBCO/BRL) (primer 1, CGGCCTTGAGCTAAGTCTATTATAAGG; primer 2, GGTGGGATTAGATAATGCCTGCTCT; primer 3, CCAAGATGACTGGTCGTACATAAG). Reaction conditions were 94°C for 5 min, 60°C for 2 min, and 72°C for 2 min (×1); 94°C for 1 min, 60°C for 2 min, and 72°C for 2 min (×30); and 94°C for 1 min, 60°C for 2 min, and 72°C for 10 min (×1). Ten microliters of product was analyzed on a 4% Tris·Borate EDTA agarose gel with the wild-type allele generating a 164-bp product and the targeted allele generating a 194-bp product.
Reagents and Administration.
Mice (8–12 weeks old) were given i.p. injections of MNNG (50 mg/kg body weight), temozolomide (100 mg/kg), cisplatin (10 mg/kg), or O6-benzylguanine (BeG) (60 mg/kg). The volume for all reagents was 0.25 ml. All drugs, except cisplatin (David Bull Laboratories, Warwick, U.K.), were prepared fresh, were first dissolved in DMSO (10% vol/vol), and were made to a final concentration with PBS (temozolomide and BeG) or corn oil (MNNG). Mice were exposed to γ-irradiation by using a 137Cs source at 0.27 Gy⋅min−1 for 15 min, so that each animal received a dose of 4 Gy.
Quantitation of Apoptosis.
At each indicated time point after injection, a minimum of three animals were killed, and the small intestine was removed, was flushed with water, and was fixed overnight in methacarn (4 parts methanol, 2 parts chloroform, 1 part acetic acid). Histological sections were made, and apoptosis was scored as described (9). A minimum of 50 half crypts were scored per animal.
Cell Culture Experiments.
ES cells were maintained under standard conditions in lymphocyte inhibitory factor-supplemented media. After electroporation with the PNT (10) vector, which contains both neomycin and herpes simplex virus thymidine kinase gene cassettes, clones were selected in 200 μg/ml G418. Clones were expanded and then were exposed to temozolomide in serum-free medium for 4 hours. Cells were subsequently maintained out of selection for a period of 4 days to permit endogenous levels of herpes simplex virus thymidine kinase to decline. Cells then were plated at a range of cell densities. Clonogenicity and mutation frequency were respectively scored by counting viable colonies after 14 days of culture in either normal medium or after selection in 2 mM ganciclovir.
RESULTS
In Vivo Mutation Frequency Scored at the Dlb-1 Locus.
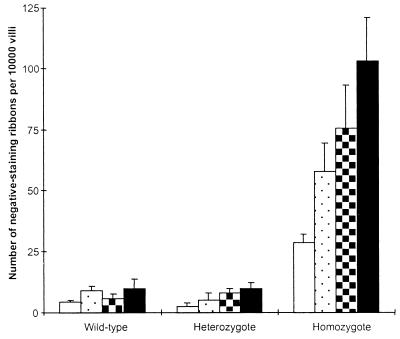
Andrew et al (11) recently demonstrated striking increases in mutation frequency in Msh2-deficient mice after treatment with N-methyl-N-nitrosourea by using a transgenic LacI reporter system. We wished to address this same question specifically within the stem cell population of the murine intestine, both at endogenous levels of DNA damage and also after treatment with the cancer chemotherapeutic drug temozolomide. The in vivo mutation frequency was scored at the Dlb-1 locus within the crypt stem cells of the intestinal epithelium of animals that were wild-type, heterozygous, or null for Msh2 and heterozygous (a/b) at the Dlb-1 locus. Twenty days after exposure to Temozolomide, Dlb-1 mutations were scored as non-lectin binding clones by using standard techniques (9, 12). Mutation frequencies in untreated and drug treated animals are shown in Fig. 1. Compared with wild-type mice, Msh2 −/− mice were found to have a significantly increased frequency of mutations both at endogenous levels of damage and after temozolomide treatment (P < 0.05, Mann–Whitney U test).
Figure 1.
Dlb-1 mutation frequencies after exposure to temozolomide. Mutation frequencies were determined from intestinal wholemounts prepared 20 days after i.p. injection with a range of doses of temozolomide. Columns represent mean mutation frequency at the Dlb-1 locus at the drug dosage shown. Error bars represent SEM. A minimum of three animals was scored per group. Open bars, 0 mg/kg; stippled bars, 25 mg/kg; hatched bars, 50 mg/kg; filled bars, 100 mg/kg. Under all conditions, Msh2 −/− mice had significantly higher mutation frequencies compared with wild-type and heterozygote animals (P < 0.05, Mann–Whitney U test).
Mutation Frequency and Clonogenicity in Vitro.
The observed increase in mutation frequency may arise directly from failed DNA repair or as a consequence of increased survival of cells bearing DNA damage. It is currently not possible to determine the clonogenic survival of individual crypt stem cells because each crypt contains multiple stem cells and in vivo clonogenicity assays rely on ablation of this entire population. We have therefore addressed this question in an in vitro model system. We determined clonogenicity and mutation frequency in ES cells wild-type or mutant for Msh2 that had been stably transfected with PNT (10), a vector encoding both the herpes simplex virus thymidine kinase gene and the neomycin phosphotransferase gene. The mutation frequency was scored at the thymidine kinase locus on the basis of resistance to the drug ganciclovir. Cultures were maintained under G418 selection to eliminate cells in which the entire vector had been inactivated: for example, after chromosome loss. After a 4-hour exposure to 2 mM temozolomide, 21% of Msh2 −/− cells formed colonies. This figure was reduced to 1.57% in +/+ cells (these figures represent the mean clonogenicity from five different experiments, each performed in triplicate and corrected for the plating efficiency of untreated cells). The mutation frequency, as scored by ganciclovir resistance, was 1.002 × 10−3 in −/− cells and 9 × 10−6 in +/+ cells (mean values derived from three different experiments performed in triplicate). These results show that Msh2 deficiency confers a 13-fold increase in clonogenicity and a 110-fold increase in mutation frequency. A significant component of the Msh2-dependent elevation in mutation frequency is therefore directly attributable to increased survival of Msh2 −/− cells.
Msh2-Dependent Apoptosis.
To test directly whether Msh2 has a significant role in the induction of apoptosis in vivo after DNA damage, we analyzed the prevalence of apoptosis after exposure to the methylating agents MNNG and temozolomide. We similarly analyzed the effect of the cross-linking agent cisplatin caused by the recognized association between cisplatin resistance and mismatch repair deficiency and also because cisplatin 1,2 intrastrand adducts may be directly recognized by the MSH2/GTBP complex (13). Apoptosis was scored within the crypts of the small intestine. This system was chosen for three principal reasons: first, the known intestinal expression of MSH2 (14); second, the demonstrated role of Msh2 in prevention of intestinal neoplasia (15); and third, because apoptosis within the murine small intestine has been well characterized (9, 16) .
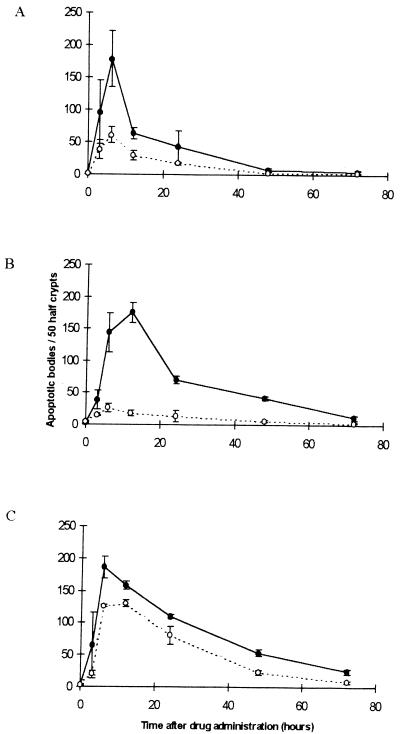
Apoptosis was scored histologically within the crypts of cohorts of mice that were either wild-type (+/+) or homozygous null (−/−) for Msh2 over a 72-hour period after drug administration (Fig. 2). In Msh2 +/+ animals, all of the drugs used induced measurable increases in intestinal apoptosis, and, in all cases, the peak induction of apoptosis was seen between 8 and 16 hours. In comparison to the pattern observed in the Msh2 +/+ animals, the apoptotic response of Msh2 −/− mice was significantly diminished for all three drug types (P < 0.05, Mann–Whitney U test). The Msh2-dependent decrease in intestinal apoptosis was greatest for temozolomide and least for cisplatin. In both Msh2 +/+ and −/− mice, apoptosis occurred at peak levels within the replicative compartment of the crypt with the mode lying between cell positions 4 and 6, overlapping the stem cell region; thus, Msh2 status influenced the number but not the spatial distribution of apoptosis within the crypt (data not shown).
Figure 2.
The in vivo apoptotic response after drug exposure. Cohorts of Msh2 +/+ or −/− animals were given i.p. injections of 50mg/kg MNNG (A); 100 mg/kg temozolomide (B); or 10mg/kg cisplatin (C) at time zero. The data show the means of the prevalence of apoptotic bodies per 50 half crypts. Each point represents data from a minimum of three mice. Closed circles, Msh2 +/+; open circles, Msh2 −/−. Error bars represent SEM. The difference between +/+ and −/− mice was significant for all three drugs at 6 and 12 hours (P < 0.05, Mann–Whitney U test). Significant differences also were seen for temozolomide-treated animals at 24 and 48 hours and for Cisplatin treated mice at 24, 48, and 72 hours.
Apoptosis Is Unaffected After BeG Administration.
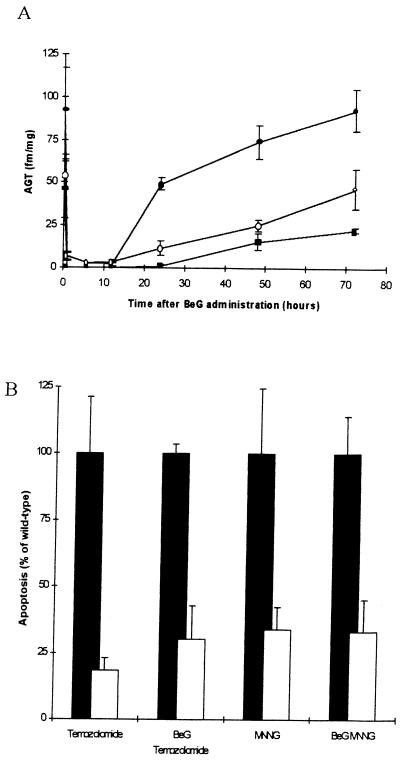
The DNA repair protein O6-alkylguanine-DNA-alkyltransferase (AGT) functions by recognizing and removing specific alkyl lesions from DNA. Failure of AGT to repair O6-methyl-G results in O6-methyl-G:T mispairs after replication, and it is postulated that these mispairs mediate cell death via mismatch repair (7). AGT activity may therefore complicate these results by effectively removing methylation damage and so suppressing cell death such that the extent of Msh2-dependent apoptosis is underestimated. To investigate this possibility, we used the inhibitor BeG to deplete AGT before challenge with MNNG and temozolomide (17). Effective depletion of AGT activity was confirmed with no detectable AGT activity between 1 and 12 hours after BeG administration (Fig. 3A). No induction of apoptosis was associated with BeG treatment alone (data not shown). Apoptosis was scored in Msh2 +/+ and −/− animals 6 hours after administration of MNNG and temozolomide and 7 hours after administration of BeG. At this time, apoptosis again was observed to depend on Msh2 status, with a significant reduction observed in Msh2 −/− mice after MNNG and temozolomide (P < 0.05, Mann–Whitney U test in both cases). The degree of Msh2 dependency was not significantly different in non-BeG treated mice (Fig. 3B).
Figure 3.
Influence of AGT on Msh2-dependent apoptosis. To competitively deplete AGT function, BeG at a dose of 60 mg/kg body weight was injected i.p. AGT levels were scored in a range of tissues as described (18). (A) Sample data is given for the liver (filled circles), colon (open circles), and small intestine (filled squares). In all three tissues, depletion of AGT was seen for the period between 1 and 12 hours after BeG injection. (B) One hour after BeG treatment, cohorts of Msh2 +/+ and −/− mice were treated with 50 mg/kg MNNG and 100 mg/kg temozolomide. Small intestines were harvested 6 hours after drug treatment, and apoptosis was scored. Data are presented showing the percentage reduction in observed apoptosis in the Msh2 −/− mice (open bars) relative to that in Msh2 +/+ mice (filled bars standardized to 100%). Non-BeG treated controls are shown for comparison. Each bar represents data from 6 mice. Error bars represent SEM.
P53 Dependency.
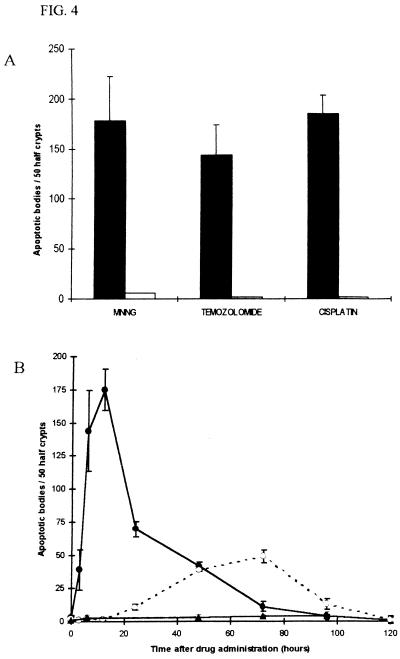
To determine whether the observed apoptosis was mediated by a p53-dependent pathway, we investigated intestinal apoptosis after drug administration in p53 −/− mice. In contrast to p53 +/+ mice, none of the drugs used induced apoptosis above background levels at 6 hours (Fig. 4A). Delayed p53-independent apoptosis was, however, observed from 24 to 96 hours after temozolomide administration in p53 −/− mice (Fig. 4B). Mice doubly mutant for both Msh2 and p53 failed to show this delayed p53-independent apoptosis (P < 0.001, Mann–Whitney U test at 72 hours) (Fig. 4B). We finally formally excluded the possibility that Msh2 may have a general role in signaling intestinal apoptosis after DNA damage. Apoptosis of lower crypt cells mediated both by p53-dependent and -independent pathways can be initiated by exposure to γ-irradiation (12, 16). However, this apoptosis is quantitatively identical in Msh2 +/+ and −/− animals (data not shown). Hence, the signal for apoptosis in lower crypt cells injured by methylating agents is specified by recruitment of Msh2.
Figure 4.
p53-dependent apoptosis in the small intestine. (A) Mice that were p53 +/+ or −/− were given i.p. injections of MNNG (50 mg/kg), temozolomide (100 mg/kg), or cisplatin (10 mg/kg) at time zero. The mean prevalence of apoptotic bodies at 6 hours is shown. Each bar represents the data from a minimum of three mice. Closed boxes, p53 +/+; open boxes p53 −/−. (B) Time course of the levels of apoptosis in p53 +/+ and −/− mice, and also mice doubly mutant for both Msh2 and p53 after injection of temozolomide (100 mg/kg) at time zero (wild-type data reproduced from Fig. 2B). Each point represents the data from a minimum of three mice. Closed circle symbols, p53 +/+; open circle symbols, p53 −/−; closed triangular symbols, Msh2 −/− and p53 −/−. Error bars represent SEM.
DISCUSSION
These data clearly show that, in response to methylating agent damage, Msh2 is required for the bulk of both the immediate and delayed apoptotic responses of lower crypt cells. Moreover, although the signal for apoptosis is processed through p53 where this is available, a death signal is generated in the absence of p53. Significantly, in the absence of both genes, no apoptosis was observed. These findings imply that one function of Msh2 is to mediate the deletion of cells bearing DNA damage of methylation type. It remains possible that the cohort of damaged cells protected from apoptosis by the absence of Msh2 may be removed by death outside the time span analyzed here or that these cells may be prevented from propagating by some other mechanism, such as permanent cell cycle arrest. Nonetheless, the phenomenon of delayed apoptosis that has been observed in p53 −/− mice after γ-irradiation (12, 16) and seen here after temozolomide was not observed in Msh2 −/− mice. Hence, the most direct conclusion from this study must be that a significant number of cells escape death by virtue of Msh2 deficiency and are therefore potential founders of clones bearing unrepaired mutations.
We also have demonstrated that functional Msh2 suppresses the mutation frequency at the Dlb-1 locus after in vivo exposure to temozolomide. This may arise either through the Msh2-dependent deletion of cells we characterize here or as a consequence of Msh2-dependent DNA repair. The in vivo experiments described here cannot discriminate between these two possibilities; however, in vitro data obtained from Msh2 −/− cells show that, in ES cells, a significant element of the increase in total mutation burden does arise from increased cell survival as assessed by increased clonogenicity. The remaining pertinent question asks, therefore, whether clonogenicity scored in ES cells is directly applicable to clonogenic survival within the intestinal epithelium. An absolute translation of these results to ES cells is questionable for a number of reasons. First, stem cells within the intestine are relatively poorly characterized, and it is probable that ES cells differ from their intestinal counterparts in a number of as yet uncharacterized ways. Second, as discussed above, in vivo clonogenic survival of intestinal stem cells is always determined at the level of entire crypts. This makes it impossible to determine the clonogenicity of individual stem cells and also to account for adaptive responses within the crypt, such as the recruitment to the stem cell population of cells normally regarded as external to this compartment. Thus, clonogenicity modeled in ES cells in vivo may not directly translate into the intestinal crypt. However, we have demonstrated the principal that, within a stem cell population, clonogenic survival modulates mutation frequency. The hypothesis that this is an important in vivo phenomenon now requires testing.
We propose two possible mechanisms to explain the involvement of p53 in engaging apoptosis. First, it has been proposed that, after methylation, cells die as a result of repeated attempts to correct the mispairs that are generated during new DNA synthesis. Such repeated abortive repair may recruit p53 and initiate apoptosis, particularly because p53 is known to bind single stranded DNA (19). Second, p53 is known to recognize insertion/deletion mismatches (20), and it has been shown that p53 forms stable complexes at these lesions that may subsequently recruit other proteins. However, we consider this latter mechanism unlikely because p53 has been shown not to bind GT mismatches.
The finding that administration of the AGT inhibitor BeG does not alter the apoptotic response is surprising in light of experiments that show that AGT inhibition normally sensitizes cells to the effects of alkylation damage (21). This suggests two possibilities: either that lesions other than O6-methyl-G may trigger Msh2-dependent apoptosis or that the drug regimes used here have already functionally depleted AGT function irrespective of the addition of BeG.
In conclusion, we have shown that a significant proportion of the normal in vivo apoptotic response of small intestinal cells to two different methylating agents and to the cross-linking agent cisplatin depends on functional Msh2 and that the degree of this reliance is strongly drug type-dependent. These observations therefore extend previous studies of methylation tolerance to an in vivo setting and identify the cellular mechanism of such tolerance. We also show that Msh2 deficiency leads to a marked increase in the in vivo mutation frequency and, furthermore, that, when modeled in vitro, a significant element of this rise occurs as a consequence of increased clonogenicity. Given that methylation damage is present in human gastrointestinal DNA (22), these findings raise the possibility that Msh2 deficiency may predispose to malignancy not only by failing to repair DNA damage but also by failing to engage apoptosis and delete damaged cells in the replicative and stem cell regions of the crypt.
These conclusions raise predictions relevant to tumorigenesis that may now be approached by using mice deficient in Msh2 and p53. For example, an Msh2-deficient background should predispose to tumor formation after exposure to temozolomide but not cisplatin. Similarly, p53 deficiency should show increased predisposition after exposure to both reagents, and deficiency of both genes should lead to a marked sensitivity to temozolomide. Such studies should allow a correlation between the ability to engage cell death and increased tumor predisposition, although the remaining challenge will be to establish the precise contributions of failed cell death and failed DNA repair to this process.
Acknowledgments
The authors thank J. Verth and staff for maintenance of the mouse colony. Temozolomide was a kind gift from M. Stevens. We thank R. S. McElhinney for the BeG. A.R.C. is a Royal Society University Research Fellow. The authors are supported by the Cancer Research Campaign, Association for International Cancer Research, and Scottish Hospital Endowment Research Trust.
ABBREVIATIONS
- MNNG
N-methyl-N′-nitro-N-nitrosoguanidine
- AGT
O6-alkylguanine-DNA-alkyltransferase
- BeG
O6-benzylguanine
- ES cell
embryonic stem cell
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Fishel R, Kolodner R D. Curr Opin Genet Dev. 1995;5:382–395. doi: 10.1016/0959-437x(95)80055-7. [DOI] [PubMed] [Google Scholar]
- 2.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 3.Nicolaides N C, Papadopoulos N, Liu B, Wei Y F, Carter K C, Ruben S M, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, et al. Nature (London) 1994;371:75–79. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 4.De Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 5.Prolla T A, Baker S M, Harris A C, Tsao E L, Yao X, Bronner C E, Zheng B H, Gordon M, Reneker J, Arnheim N, et al. Nat Genet. 1998;18:276–279. doi: 10.1038/ng0398-276. [DOI] [PubMed] [Google Scholar]
- 6.Drummond J T, Li G, Longley M J, Modrich P. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 7.Karran P, Bignami M. BioEssays. 1994;16:833–839. doi: 10.1002/bies.950161110. [DOI] [PubMed] [Google Scholar]
- 8.Winton D J, Blount M A, Ponder B A. Nature (London) 1988;333:463–466. doi: 10.1038/333463a0. [DOI] [PubMed] [Google Scholar]
- 9.Clarke A R, Gledhill S, Hooper M L, Bird C C, Wyllie A H. Oncogene. 1994;9:1767–1773. [PubMed] [Google Scholar]
- 10.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 11.Andrew S E, McKinnon M, Cheng B S, Francis A, Penney J, Reitmair A H, Mak T W, Jirik F R. Proc Natl Acad Sci USA. 1998;95:1126–1130. doi: 10.1073/pnas.95.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke A R, Howard L A, Harrison D J, Winton D J. Oncogene. 1997;14:2015–2018. doi: 10.1038/sj.onc.1201040. [DOI] [PubMed] [Google Scholar]
- 13.Yamada M, O’Regan E, Brown R, Karran P. Nucleic Acids Res. 1997;25:491–495. doi: 10.1093/nar/25.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leach F S, Polyak K, Burrell M, Johnson K A, Hill D, Dunlop M G, Wyllie A H, Peltomaki P, de la Chapelle A, Hamilton S R, et al. Cancer Res. 1996;56:235–240. [PubMed] [Google Scholar]
- 15.DeWind N, Dekker M, VanRossum A, VanderValk M, Riele H T. Cancer Res. 1998;58:248–255. [Google Scholar]
- 16.Merritt A J, Allen T D, Potten C S, Hickman J A. Oncogene. 1997;14:2759–2766. doi: 10.1038/sj.onc.1201126. [DOI] [PubMed] [Google Scholar]
- 17.Dolan M E, Moschel R, Pegg A E. Proc Natl Acad Sci USA. 1990;87:5368–5372. doi: 10.1073/pnas.87.14.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rafferty J A, Clarke A R, Sellappan D, Koref M S, Frayling I M, Margison G P. Oncogene. 1996;12:693–697. [PubMed] [Google Scholar]
- 19.Wu L, Bayle J H, Elenbaas B, Pavletich N P, Levine A J. Mol Cell Biol. 1995;15:497–504. doi: 10.1128/mcb.15.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S, Elenbaas B, Levine A, Griffith J. Cell. 1995;81:1013–1020. doi: 10.1016/s0092-8674(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 21.Pegg A E, Swenn K, Chae M Y, Dolan M E, Moschel R C. Biochem Pharmacol. 1995;50:1141–1148. doi: 10.1016/0006-2952(95)00249-y. [DOI] [PubMed] [Google Scholar]
- 22.Hall C N, Badawi A F, O’Connor P J, Saffhill R. Br J Cancer. 1991;64:59–63. doi: 10.1038/bjc.1991.239. [DOI] [PMC free article] [PubMed] [Google Scholar]