Abstract
Amyloid β peptide (Aβ), the principal proteinaceous component of amyloid plaques in brains of Alzheimer’s disease patients, is derived by proteolytic cleavage of the amyloid precursor protein (APP). Proteolytic cleavage of APP by a putative α-secretase within the Aβ sequence precludes the formation of the amyloidogenic peptides and leads to the release of soluble APPsα into the medium. By overexpression of a disintegrin and metalloprotease (ADAM), classified as ADAM 10, in HEK 293 cells, basal and protein kinase C-stimulated α-secretase activity was increased severalfold. The proteolytically activated form of ADAM 10 was localized by cell surface biotinylation in the plasma membrane, but the majority of the proenzyme was found in the Golgi. These results support the view that APP is cleaved both at the cell surface and along the secretory pathway. Endogenous α-secretase activity was inhibited by a dominant negative form of ADAM 10 with a point mutation in the zinc binding site. Studies with purified ADAM 10 and Aβ fragments confirm the correct α-secretase cleavage site and demonstrate a dependence on the substrate’s conformation. Our results provide evidence that ADAM 10 has α-secretase activity and many properties expected for the proteolytic processing of APP. Increases of its expression and activity might be beneficial for the treatment of Alzheimer’s disease.
The amyloid precursor protein (APP) is a type I transmembrane glycoprotein constitutively expressed in many types of mammalian cells. APP is the precursor of the amyloid β peptide (Aβ), the principal proteinaceous component of amyloid plaques in brains of Alzheimer′s disease patients (1, 2). The Aβ sequence includes 28 amino acids of the extracellular and 12–15 residues of the membrane-spanning region of APP. Aβ is derived by proteolytic processing of the precursor protein from as yet not identified proteases, the β-secretase cleaving at the N terminus, and the γ-secretase cleaving at the C terminus.
The major proteolytic pathway of APP is the constitutive secretory pathway that involves cleavage by a putative α-secretase within the Aβ sequence at the cell surface (3–5) and in the trans-Golgi network (6–9). Soluble N-terminal APP (APPsα) fragments of 105–125 kDa (10) are released into the extracellular medium. The membrane-bound 10-kDa C-terminal fragment (p10) produced by α-secretase cleavage of APP contains only part of the amyloidogenic Aβ and can be further cleaved by the γ-secretase to yield a secreted 3-kDa fragment (p3) (11). Because this pathway does not produce intact Aβ, it is nonamyloidogenic and cannot lead to Alzheimer’s disease pathology.
There has been intensive interest in the secretase enzymes that cleave APP in relation to the pathology of Alzheimer′s disease. The putative α-secretase cleavage site has been precisely determined (12): in human embryonic kidney cells (HEK 293) transfected with various constructs of APP, the soluble form ends at Gln-15 of Aβ, and the N terminus of the cleaved C-terminal fragment begins at Leu-17. It was proposed that Lys-16 was split from either fragment by an ectopeptidase. Sequence analysis of secreted human brain APP showed only one product ending at Gln-15 (13), whereby a carboxypeptidase release of Lys-16 could not be ruled out. Studies in various cell types confirmed that the major α-secretory cleavage site is after Lys-16 in the Aβ domain, but multiple minor cleavages around this site have been observed (14, 15). Principal determinants of APP cleavage by α-secretase appear to be the distance of the hydrolyzed bond from the membrane (12 or 13 residues) and a local helical conformation (3).
Besides APP, many other transmembrane proteins can undergo proteolytic cleavage and release of their extracellular domain. This proteolytic process is often referred to as “ectodomain shedding” and affects cell surface molecules such as growth factors, growth factor receptors, ectoenzymes, and cell adhesion molecules. Recently a metalloproteinase-disintegrin protein has been identified (16, 17) that specifically cleaves precursor tumor necrosis factor α (TNF-α) and releases TNF-α from cells. This enzyme, called TNF-α converting enzyme or TACE, belongs to a family of membrane-anchored glycoproteins that are composed of several distinct protein domains including a pro- and metalloprotease domain, a disintegrin domain, a cysteine-rich region, and an epidermal growth factor repeat. Members of the ADAM family (ADAM stands for a disintegrin and metalloprotease) have been implicated in several cell interactive events including cell–cell fusion (18, 19). In Drosophila melanogaster neurogenesis, the metalloprotease disintegrin protein KUZ is required in the early embryo for neural inhibition (20). There is evidence that this process mediated by KUZ involves a specific cleavage in the extracellular domain of the transmembrane receptor Notch (21) and of the membrane-bound Notch ligand Delta (22).
We isolated a membrane-bound metalloprotease from bovine kidney that was identified as ADAM 10, a member of the metalloprotease disintegrin protein family. ADAM 10 and KUZ share a high level of sequence similarity throughout the molecule (41% amino acid identity). The enzyme has been first purified (23) and cloned (24) from bovine brain and can digest myelin basic protein in vitro, though it is not clear whether myelin basic protein is a natural substrate in vivo. During our studies with ADAM 10, we noticed a striking similarity between the inhibition of ADAM 10 by various inhibitors and the results reported for inhibition of APP cleavage by a putative α-secretase (25). We therefore investigated whether ADAM 10 has properties expected for a physiologically relevant APP-processing enzyme with α-secretase activity.
MATERIALS AND METHODS
Materials.
Peptides were prepared by a solid-phase method using fluorenylmethoxycarbonyl chemistry on the model 9050 Plus Millipore peptide synthesizer. The hydroxamic acid-based inhibitor BB-3103 was provided by British Biotech (Oxford, U.K.). We used the following antibodies: anti-hemagglutinin (HA) IgG (mouse monoclonal antibody, clone 16B12, Babco, Richmond, CA; rabbit polyclonal antibody, Y-11, Santa Cruz Biotechnology), anti-rabbit Ig horseradish perioxidase-linked whole antibody, and 35S-labeled anti-mouse IgG whole antibody (Amersham). Antibodies against APP were as follows: mouse monoclonal IgG 6E10 from Senetek (St. Louis), and rabbit polyclonal antibodies 1736 and C7 were provided by D. J. Selkoe (Harvard Medical School, Boston).
Purification of ADAM 10 Protease.
ADAM 10 protease was purified to homogeneity from bovine kidney plasma membranes by modifying the method of Howard and Glynn (26). Analysis of the protein fractions containing enzyme activity by SDS/PAGE showed a protein with a molecular weight of about 62,000. N-terminal sequence analysis revealed the sequence TTVXEKNTXQLYIQTDXXFF, which is identical to a sequence of bovine MADM (24) also known as ADAM 10.
Protease Assay for Determination of Substrate Specificity.
Peptides in a final concentration of 0.3 mM were incubated with 2 μg of purified enzyme in 50 μl containing 50 mM N-2-hydroxyethylpiperazine-N′-(3-propanesulfonic acid) (pH 8.3) and 250 mM NaCl at 37°C for 30 min or 6 hr. Reactions were stopped with 2 μl of trifluoroacetic acid (TFA) and samples were analyzed by HPLC on a Vydac RP 18 column. Peptides were separated with the buffer system 0.1% TFA in water (buffer A) and 0.1% TFA, 9.9% water, and 90% acetonitrile (buffer B) by using the following gradients: 0% B, 5 min; 0–70% B, over 45 min. For calculation of percent cleavage values, the peak area of peptide incubated without enzyme and treated identically was used as reference for 100%.
CD Spectroscopy.
CD measurements were carried out on a Jasco J720 spectropolarimeter at 4°C. The path length of the quartz cell was 0.1 mm. The measurements were performed at a peptide concentration of 0.5 mM in 20 mM Tris⋅HCl, pH 8.3/250 mM NaCl/0.5% SDS. Results are expressed in terms of ellipticity [Θ] (degrees × 10−3).
Cloning and Functional Expression of ADAM 10.
PCR primers corresponding to the nucleotide sequence of accession no. Z21961 (EMBL Data Bank) were used to amplify the cDNA of bovine ADAM 10 from a bovine kidney cDNA library. The cDNA of ADAM 10 was fused with a synthetic DNA cassette coding for the HA-epitope tag (YPYDVPDYA). Comparison of the cloned nucleotide sequences coding for bovine ADAM 10 with the originally published sequence (24) revealed three nucleotide exchanges: C700T, C2122T, and C2251T (the nucleotide coordinates are from accession no. Z21961). The substitutions do not alter the translated protein.
For expression of C-terminal HA-tagged ADAM 10, the expression vector pcDNA3 (Invitrogen) was used. Stable transfection of HEK 293 cells was performed by the calcium phosphate precipitation technique. For selection of transfected cells, Geneticin (1 mg/ml) was used. Overexpression of ADAM 10 did not increase the growth rate of transfected HEK cells.
To generate the dominant negative form of ADAM 10, the amino acid substitution Glu → Ala at position 384 (E384A) was introduced. Mutated ADAM 10 cDNA was cloned into pcDNA3 adding a DNA cassette coding for a FLAG epitope tag (DYKDDDDK) at the 3′ end at the same time. HEK cells were transfected and selected as described above. The expression of mutated ADAM 10 was detected by Western blot analysis.
Reverse Transcriptase–PCR.
Total RNA from Madin–Darby bovine kidney (MDBK) cells and HEK cells was prepared with the RNeasy kit (Qiagen, Hilden, Germany). For reverse transcription of human and bovine ADAM 10 mRNA, a completely matching oligonucleotide for the 3′ ends of the mRNAs was used (5′-TGCACCGCATGAAAACATC-3′). PCR was performed by adding the forward primer 5′-GAACAAGGTGAAGAATGTG-3′ with sequence identity between human and bovine ADAM 10 cDNA.
Expression and Deglycosylation of ADAM 10.
All cells were grown in DMEM supplemented with 10% FBS, penicillin (100 units/ml), streptomycin (100 μg/ml), and 2 mM glutamine in 10-cm dishes to confluency. Cells were lysed in 1 ml of lysis buffer [20 mM imidazole, pH 6.8/0.2% Triton X-100/100 mM KCl/2 mM MgCl2/10 mM EGTA/300 mM sucrose/BSA (1 mg/ml)] in the presence of inhibitors (complete mini, Roche) for 20 min on ice. The clarified extracts were incubated with anti-HA antibody (16B12; 6.6 μg/ml) and protein-A Sepharose at 4°C overnight. For deglycosylation experiments, immune complexes were boiled in 100 μl of 1% SDS/2% 2-mercaptoethanol for 10 min. After centrifugation, the eluate was incubated with 2,500 units of N-glycosidase F in 50 mM sodium phosphate, pH 7.5/1.5% NP-40/0.25% SDS for 3 hr at 37°C. Proteins were precipitated as described (27), separated on a 4–12% NuPAGE gel (Novex), and blotted onto poly(vinylidene difluoride) (PVDF) membranes. Membranes were probed with anti-HA antibody (Y-11) at a dilution of 1:1,000, followed by an anti-rabbit horseradish peroxidase coupled antibody. Detection was performed by using the ECL system (Amersham).
Cell Surface Biotinylation.
Cells were grown to 90% confluency and incubated with PBS containing sulfo-N-hydroxy–succinimidobiotin (0.3 mg/ml) for 30 min at 4°C. Excess biotinylating reagent was quenched by washing several times with TBS. Cell lysates were immunoprecipitated with anti-HA mAb as described above. Immune complexes were eluted from washed protein A-beads with 200 μl of 100 mM glycine, pH 2.5/20 μM competing HA peptide (Roche). Supernatants were collected, neutralized with 5 μl of 3 M Tris (pH 8.9), and incubated with streptavidin-agarose beads for 2 hr at 4°C. Samples were analyzed by SDS/PAGE on 10% gels and blotted onto PVDF membranes. Blots were analyzed as described above.
Immunoblot Analysis.
Approximately 8 × 105 cells were seeded and grown close to confluency on 5-cm dishes treated with poly-(l-lysine). They were incubated in the absence or presence of the indicated compound for 4 hr in serum-free DMEM containing fatty acid-free BSA (10 μg/ml). Medium was collected and proteins were precipitated with 10% (vol/vol) trichloroacetic acid. Samples were centrifuged at 20,800 × g and pellets were washed twice with 500 μl of ice-cold acetone. Proteins were separated by SDS/PAGE in 7.5% gels and blotted onto PVDF membranes. Membranes were probed with antibody 6E10 at a dilution of 1:2,000 followed by a 35S-labeled anti-mouse IgG antibody. The radioactive bands corresponding to APPsα were quantified with the Bio-Imaging Analyzer BAS-1800 (Fuji). The protein content of each cell culture dish was determined after lysis with the Micro-BCA protein assay (Pierce), and the values of the radioactive bands were normalized to the protein amount.
Metabolic Labeling and Immunoprecipitation.
Approximately 1.2 × 106 cells were cultured as described above on 10-cm dishes. Nearly confluent cell cultures were incubated in serum-free and cysteine/methionine-free DMEM for 1 hr, and then labeled with Tran[35S]-label (ICN; 0.2 mCi/ml; 1 Ci = 37 GBq) for 5 hr. Immunoprecipitations were carried out as described (28, 29). APPsα was immunoprecipitated with the α-secretase-specific antibody 1736 and the 10-kDa fragment was immunoprecipitated with the antibody C7. Radioactivity was detected with the Bio-Imaging analyzer model BAS-1800 (Fuji).
Confocal Microscopy.
HEK ADAM 10 cells on fibronectin-coated glass coverslips were fixed and permeabilized with 95% methanol/5% acetic acid for 5 min at −70°C. Anti-HA (Y-11) was used in a final concentration of 2 μg/ml. The Golgi marker anti-Golgi 58K (ascites fluid, Sigma) was diluted 1:100. The fluorochrome-labeled secondary antibodies (Jackson ImmunoResearch) were applied for 30 min at room temperature at a concentration of 15 μg/ml. The labeled secondary antibodies used were Texas Red-conjugated anti-mouse (for anti-Golgi 58K) and fluorescein isothiocyanate-coupled anti-rabbit (for Y-11). Confocal microscopy was carried out as described (30).
RESULTS
Specificity of Purified ADAM 10 for Peptide Substrates.
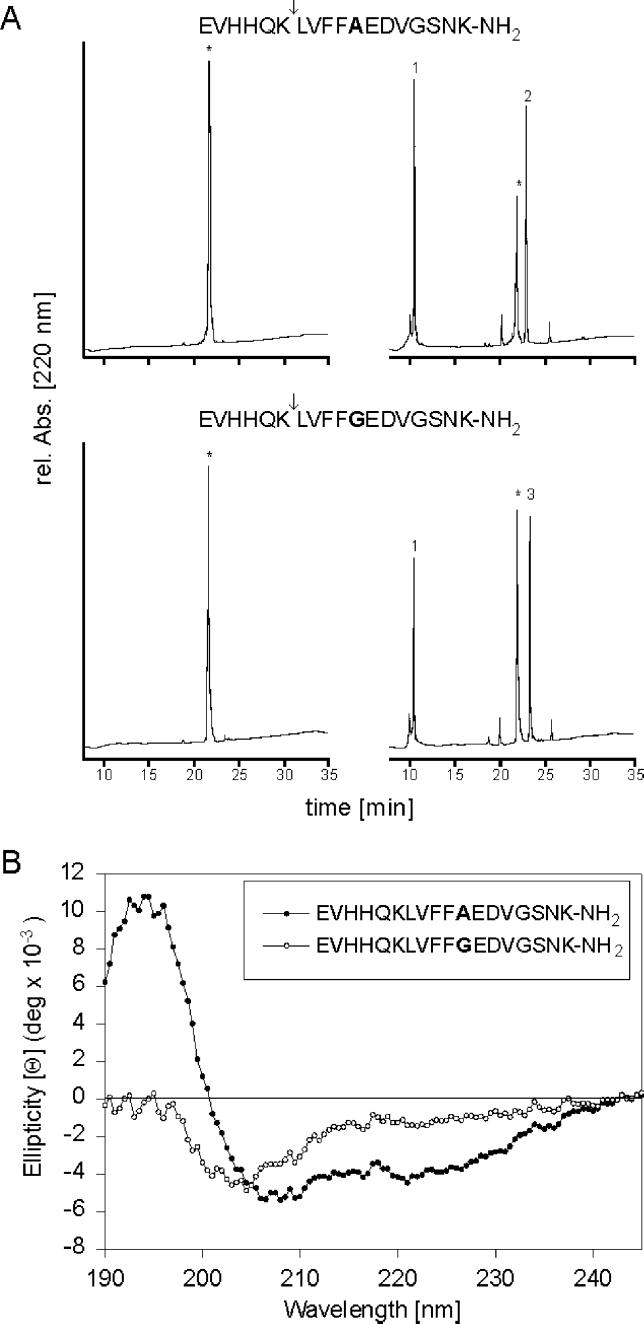
As a peptide substrate spanning the α-secretase cleaving site, we chose the octadecapeptide amide sequence, residues 11–28 in Aβ (numbering with respect of the N terminus of Aβ) (12). Its C terminus corresponds to the first extracellular residue of APP. After a 30-min incubation of Aβ(11–28) with ADAM 10, HPLC analysis showed cleavage of 28% of the starting peptide with two fragments arising (Table 1). Analysis by electrospray ionization mass spectrometry identified them as the N-terminal hexapeptide ([M+H]+ ion at m/z of 777.5) and the C-terminal dodecapeptide amide ([M+H]+ ion at m/z of 2082.8) of the parent octadecapeptide amide. Thus ADAM 10 proteolytically cleaves between Lys-16 and Leu-17 as expected for an enzyme with α-secretase activity. After a 6-hr incubation, 69% of Aβ(11–28) were cleaved predominantly at this site with minor cleavage occurring after Leu-17 and Val-18 (Fig. 1A and Table 1).
Table 1.
Specificity of purified ADAM 10 from bovine kidney for peptides spanning the cleavage site of shed proteins
| Protein | Sequence | % cleavage by bovine ADAM 10
|
|
|---|---|---|---|
| 30 min | 6 hr | ||
| APP | EVHHQK ↓ LVFFAEDVGSNK-NH2 | 28 | 69 |
| APP (A692G) | EVHHQK ↓ LVFFGEDVGSNK-NH2 | 14 | 54 |
| IL-6 receptor | ANATSLPVQ ↓ DSSSV-NH2 | 0 | 0 |
| Angiotensin-converting enzyme | TPNSAR ↓ SEGPLPDSGR-NH2 | 0 | 0 |
| Pro-TNF-α | PLAQA ↓ VRSSSRTPSD-NH2 | 49 | 100 |
Cleavage of peptides by ADAM 10 after 30 min or 6 hr was determined by HPLC analysis. ↓, proteolytic cleavage site.
Figure 1.
Cleavage of peptides spanning the α-secretase cleavage site of APP by ADAM 10. (A) Peptide substrates were incubated in cleavage buffer in the absence (Left) or in the presence of ADAM 10 (Right) for 6 hr at 37°C, followed by HPLC analysis. The asterisks indicate the peptide substrates and the numbers indicate the products generated as follows: 1, EVHHQK-OH; 2, LVFFAEDVGSNK-NH2; 3, LVFFGEDVGSNK-NH2. (B) CD spectra of APP peptides. Measurements were carried out in the presence of 0.5% SDS.
Next we examined whether the extent of cleavage depends on the conformation of the substrate as has been described for the cleavage of APP by α-secretase (3). For this purpose, Ala-21 in Aβ(11–28) was replaced by Gly. This position corresponds to a naturally occurring Ala → Gly mutation at position 692 of APP770 (31), which was identified in patients with cerebral hemorrhages due to amyloid angiopathy. In vivo studies with APP770 (A692G) showed that this mutation C-terminal to the α-secretase cleavage site led to relatively more Aβ compared with p3 by partial inhibition of α-secretase (32) and to an increased alternative cleavage of the Aβ domain, probably caused by aberrant substrate recognition of α-secretase (33).
CD measurements for the octadecapeptide amide Aβ(11–28) in 0.5% SDS showed a spectrum characteristic for α-helical conformation, whereas a random coil conformation was observed for the peptide with the Ala → Gly mutation in position 21 of Aβ(11–28) (Fig. 1B). The latter peptide was cleaved by ADAM 10 less efficiently than the wild-type peptide: after 30 min, only 14% of the peptide was cleaved between Lys-16 and Leu-17 compared with 28% of Aβ(11–28) (Table 1). ADAM 10 did not cleave peptide substrates containing a cleavage site for ectodomain shedding of angiotensin-converting enzyme (34) or IL-6 receptor (35), but it cleaved a peptide substrate derived from pro-TNF-α, as reported (36, 37).
Several metalloproteinase inhibitors were examined for their ability to inhibit cleavage of Aβ(11–28) by purified ADAM 10. The hydroxamic acid-based inhibitor BB-3103 (38) at 100 μM completely inhibited the cleaving activity. ADAM 10 was also completely inhibited by 1 mM DTT, which suggests the existence of thiol or disulfide bonds important for α-secretase activity, and by 1 mM of the chelating agent 1,10-phenanthrolin.
Processing and Localization of ADAM 10 in HEK Cells.
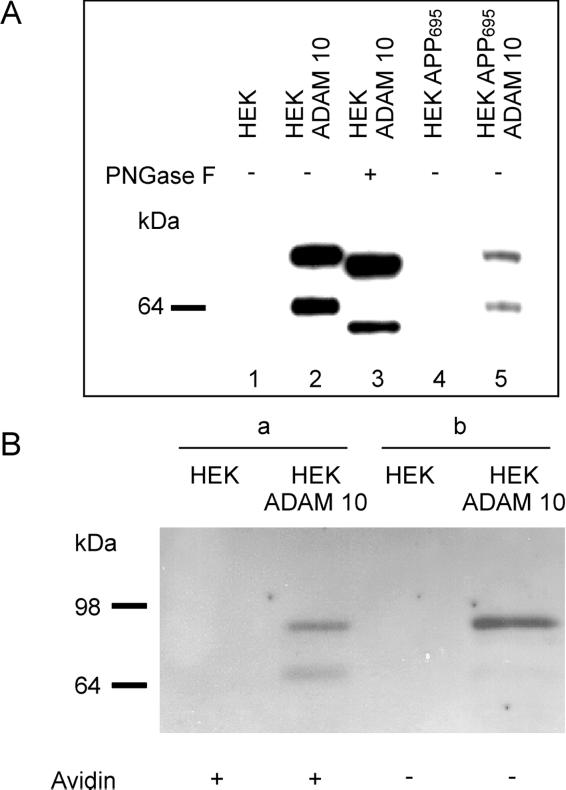
To study the effect of ADAM 10 on APP cleavage in a cellular system, we used HEK cells. The ADAM 10 cDNA was cloned from a bovine kidney cDNA library, tagged at its 3′ end with a DNA sequence coding for the HA antigen, and cloned into the expression vector pcDNA3. After transfection of HEK cells and HEK cells stably expressing APP695 (HEK APP695) (29) with ADAM 10, clones were analyzed for expression and processing of the metalloprotease. For this purpose, cell lysates were immunoprecipitated with anti-HA antibody (16B12) and analyzed by SDS/PAGE and Western blot. Immunostaining revealed two immunoreactive species with apparent molecular masses of 90 kDa and 64 kDa (Fig. 2A, lanes 2 and 5). The 64-kDa form is derived from the 90-kDa form by removal of the prodomain (194 aa), probably by a furin-like pro-protein convertase, which cleaves ADAMs at the sequence motif RXKR in a late Golgi compartment (39). The calculated molecular mass of the protein core of ADAM 10 after cleavage of the prodomain is 61 kDa. To investigate whether the difference between apparent and calculated molecular masses is caused by glycosylation on the putative N-glycosylation sites of ADAM 10 (24), the glycoproteins of the cell clone with the highest expression level of ADAM 10 (HEK ADAM 10) were treated with N-glycosidase F. This treatment resulted in a reduction of the molecular masses to the expected values of about 86 kDa for the precursor protein and about 61 kDa for the protein core of ADAM 10 lacking the prodomain (Fig. 2A, lane 3).
Figure 2.
Expression and deglycosylation of ADAM 10 protease (A). After transfection of HEK and HEK APP695 cells with ADAM 10 cDNA, cell lysates were immunoprecipitated with anti-HA antibody, subjected to deglycosylation with N-glycosidase F (PNGase F) (lane 3), or directly analyzed by 4–12% NuPAGE gel system and Western blot. (B) HEK and HEK ADAM 10 cells were surface-biotinylated. After immunoprecipitation with anti-HA antibody (16B12) and elution of the immune complexes, four-fifths of the eluate were incubated with immobilized streptavidin (lanes a), and the remaining one-fifth was directly applied to a 10% SDS/PAGE gel (lanes b) and then blotted onto PVDF membranes.
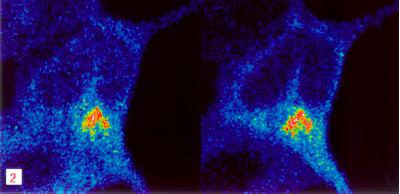
The localization of ADAM 10 in transfected HEK cells was investigated by cell surface biotinylation and confocal microscopy. For biotinylation, cells were cooled to 4°C and incubated for 30 min with sulfo-N-hydroxysuccinimidobiotin, a membrane-impermeant biotinylation reagent. Immunoprecipitation was performed with the anti-HA mAb. Four-fifths of collected immunoprecipitates were incubated with immobilized streptavidin to recover protease molecules localized on the plasma membrane. Fig. 2B shows the appearance of both the processed ∼64-kDa form of ADAM 10, as well as its 90-kDa precursor at the cell surface (lanes a). One-fifth of the immunoprecipitate that was not incubated with streptavidin predominantly showed the nonprocessed 90-kDa form and only a faint band of proteolytically activated ADAM 10 (lanes b). These results demonstrate that the proteolytically activated form of ADAM 10 is localized mainly on the cell surface, where it is able to cleave APP. The majority of ADAM 10 was found intracellularly as proenzyme. Analysis of the localization of ADAM 10 by confocal microscopy of permeabilized HEK ADAM 10 cells showed that its staining pattern largely overlapped with that of the Golgi marker 58K (Fig. 3).
Figure 3.
Colocalization of ADAM 10 and of a Golgi marker in HEK cells. Stably expressed ADAM 10 and the Golgi network were simultaneously immunostained. (Left) Anti-Golgi 58K protein. (Right) Anti-HA-epitope staining. The high degree of overlap of both immunoreactivities in the perinuclear region reveals a strong colocalization of ADAM 10 and the trans-Golgi marker. The scale bar is in μm.
Effect of ADAM 10 on α Secretase Cleavage of APP in HEK Cells.
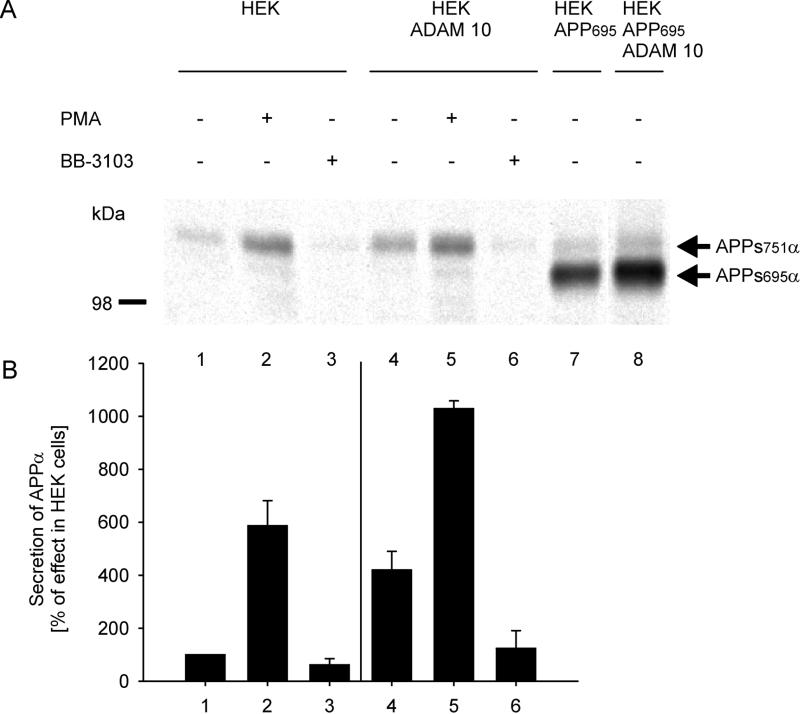
The α-secretase activity in HEK ADAM 10 cells was compared with the activity found in untransfected HEK cells. The release of APPs into the medium from cells was monitored with two site-specific antibodies, 1736 and 6E10, both of which recognize the N-terminal sequence of Aβ and thus only detect APPsα, and not APPsβ. Immunoblot experiments and immunoprecipitation after metabolic labeling with 35S were performed to determine α-secretase activity (Figs. 4 and 5). Quantitative analysis of immunoblot experiments with mAb 6E10 showed that HEK cells stably expressing a high level of ADAM 10 release approximately four times more APPsα into the medium compared with untransfected HEK cells (Fig. 4, lanes 1 and 4). Increased α-secretase activity was also observed in the HEK APP695 cell clone stably expressing ADAM 10. The relative increase of APPs751α derived from endogenous APP and of APPs695α was only 2-fold (Fig. 4A, lanes 7 and 8), because of the lower expression level of ADAM 10 (Fig. 2, lane 5) and the higher substrate to enzyme ratio.
Figure 4.
Secretion of APPsα from HEK and HEK ADAM 10 cells. (A) Cells were incubated in the presence of indicated compound. After 4 hr, the medium was collected, and proteins were precipitated and subjected to immunoblot analysis with antibody 6E10 followed by a 35S-labeled anti-mouse IgG antibody. (B) Quantitative analysis of secreted APPsα. The radioactive bands corresponding to APPsα were quantified with the Bio-Imaging analyzer model BAS-1800. The results are expressed as percentage of secreted APPsα in control HEK cells and are the averages ±SD of at least three experiments.
Figure 5.
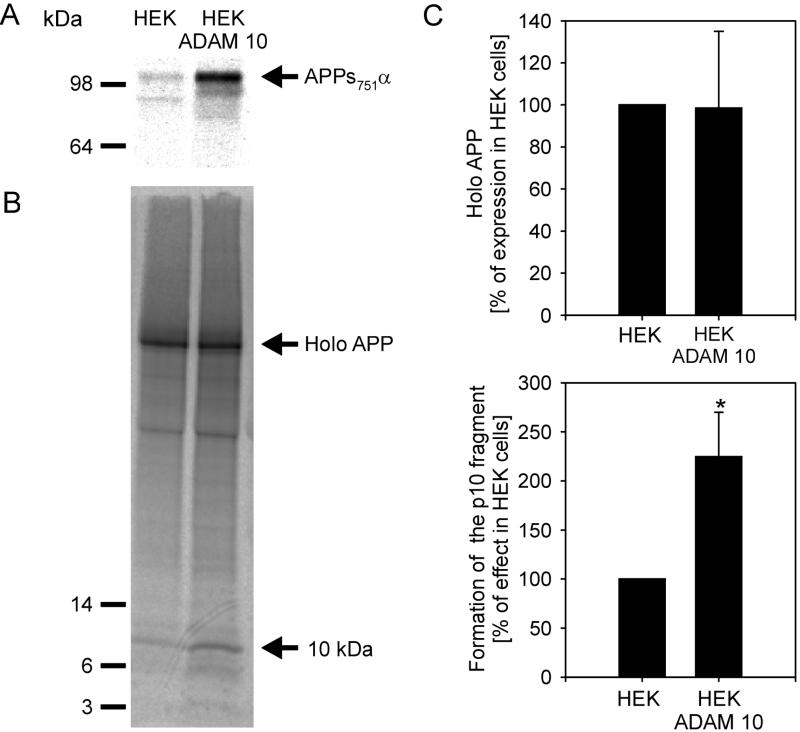
Effect of ADAM 10 on the production of APPsα and on the 10-kDa C-terminal fragment. (A) Cells were metabolically labeled with l-[35S]methionine and [35S]cysteine (200 μCi/ml) for 5 hr. Cell media were immunoprecipitated with antibody 1736 and analyzed by SDS/PAGE in 10% gels. (B) Cell lysates were immunoprecipitated with antibody C7. The samples were separated by 10–20% Tris/Tricine gel (Novex) and analyzed as described. (C) Quantitative analysis of holo-APP and p10. The values of p10 were normalized to the levels of holo-APP. The results are the averages ±SD of four experiments. Statistical significance between control cells and HEK ADAM 10 cells was determined by Student’s unpaired t test (∗, P < 0.005).
Hydroxamic acid-based zinc metalloprotease inhibitors have recently been shown to inhibit the ectodomain shedding of several membrane proteins including the α-secretase cleavage of APP. No effect on the release of APPsβ has been found (40). We therefore investigated the effect of the hydroxamic acid-based inhibitor BB-3103 on HEK cells and on HEK cells overexpressing ADAM 10. BB-3103 inhibited the release of APPsα in HEK cells by about 40% (Fig. 4, lanes 1 and 3) and in HEK ADAM 10 cells by about 70% (Fig. 4, lanes 4 and 6).
The stimulation of protein kinase C by phorbol esters strongly enhances the release of APPsα and inhibits the secretion of Aβ (41, 42). To examine the effect of protein kinase C stimulation on ADAM 10 activity, HEK and HEK ADAM 10 cells were treated with 1 μM phorbol 12-myristrate 13-acetate (PMA). Immunoblot experiments showed that PMA increased APPsα release from HEK cells about 6.0-fold (Fig. 4, lanes 1 and 2). The augmented α-secretase activity from HEK ADAM 10 cells was further increased 2.5-fold (Fig. 4, lanes 4 and 5).
Similar results were observed in immunoprecipitation experiments with antibody 1736, thus the expression of ADAM 10 in HEK cells significantly increased APPsα secretion into the medium (Fig. 5A). To determine whether ADAM 10 expression also increases the level of p10, cell lysates from HEK and HEK ADAM 10 cells after metabolic labeling were immunoprecipitated with antibody C7 recognizing the C-terminal part of APP. As shown in Fig. 5 B and C, the expression of full-length APP (holo-APP) is not changed, whereas the signal for p10 is significantly stronger in HEK ADAM 10 cells than in control cells, further indicating that ADAM 10 has α-secretase activity.
Effect of a Dominant Negative ADAM 10 Mutant on α Secretase Activity.
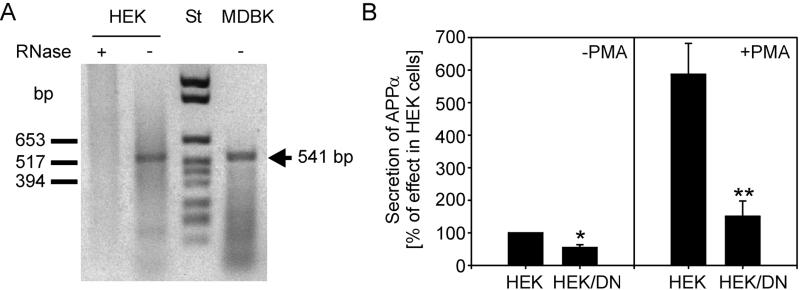
To determine whether the human homologue of ADAM 10 is expressed in HEK cells and, therefore, might be responsible for the endogenous α-secretase activity, we performed reverse transcriptase–PCR analysis using total RNA obtained from these cells and oligonucleotides specific for ADAM 10. As shown in Fig. 6A, HEK cells express detectable amounts of mRNA encoding for human ADAM 10, which shows 97% identity of amino acids with bovine ADAM 10. In a control experiment, bovine ADAM 10 mRNA was detected in total RNA of MDBK cells known to express ADAM 10 (24).
Figure 6.
Inhibition effect of a dominant negative ADAM 10 protease form (DN). (A) Endogenous expression of ADAM 10 in HEK and MDBK cells detected by reverse transcriptase–PCR. In each case a 541-bp DNA fragment could be amplified. As control, RNA from HEK cells was treated with RNase A before reverse transcription (St, DNA molecular weight marker). (B) Quantitative analysis of secreted APPsα after immunoblot analysis. The results are expressed as percentage of secreted APPsα in control HEK cells and are the averages ±SD of at least three experiments. Statistical significance between control cells and HEK/DN cells treated or untreated with PMA was determined by Student’s unpaired t test (∗, P < 0.005; ∗∗, P < 0.001).
To inhibit the endogenous α-secretase activity in HEK cells, the point mutation E384A was introduced into the zinc binding site of the bovine ADAM 10 protease. A mutant KUZ protein with a mutation at the same site acted as a dominant negative form in D. melanogaster (21). HEK cells stably expressing the mutant ADAM 10 E384A (HEK/DN) showed a substantially decreased secretion of APPs. The inhibitory effect was most apparent in PMA-treated cells: only about 25% of enzymatic activity was observed (Fig. 6B). Thus, this point mutation resulted in a dominant negative form of the protease and significantly decreased constitutive and stimulated α-secretase activity.
DISCUSSION
Despite the biomedical interest in the proteolytic cleavage of APP, the proteases involved have remained elusive. In this study, we provide evidence that the disintegrin metalloprotease ADAM 10, a type I membrane protein, has α-secretase activity and is involved in basal and stimulated ectodomain shedding of APP. These conclusions are supported by several experimental results:
(i) Overexpression of ADAM 10 in HEK cells leads to a severalfold increase of APPsα and of the p10 fragment. The increase in shedding activity is stronger in cells with a higher expression level of ADAM 10. This enhanced α-secretase activity caused by overexpression of ADAM 10 can be further increased by stimulation of protein kinase C with phorbol esters, a characteristic feature of the proposed α-secretase (41, 42). Because subtle differences in the way APP is processed were observed in primary neurons compared with other cell types (15, 43, 44), further studies including neurons are necessary to see whether our results can be generalized.
(ii) Cell surface biotinylation experiments demonstrate that the proteolytically activated form of ADAM 10 is localized mainly in the plasma membrane. This result supports the view that cleavage of the transmembrane protein APP occurs by a membrane-bound endoprotease at the cell surface (3–5). On the other hand several reports provided evidence that APP is cleaved by the α-secretase in a trans-Golgi compartment of the secretion pathway (6–9). ADAM 10 is predominantly found as proenzyme intracellularly in the Golgi, presumably in an inactive form. APP may also be cleaved along the secretory pathway from the Golgi to the plasma membrane after processing and activating ADAM 10 by a furin-like pro-protein convertase in the lumen of the secretory pathway. A significant amount of furin has been shown to cycle between the Golgi and the plasma membrane (45). Our results suggest that α-secretase cleavage can occur at the cell surface and intracellularly. The relative contribution of both probably depends on the cell type and the rate of cycling furin.
(iii) The endogenous basal and phorbol ester stimulated α-secretase activity in HEK cells can be substantially inhibited by a dominant negative form of ADAM 10, where a conserved glutamic acid residue in the zinc binding motif was mutated.
(iv) The inhibitor spectrum both of the isolated and the overexpressed enzyme is consistent with the reports for inhibition of α-secretase activity. Furthermore, cleavage of Aβ peptide fragments by ADAM 10 requires a minimum length and is conformation-dependent. Therefore, an octapeptide spanning the α-secretase cleaving site was not a suitable substrate for human ADAM 10 (37). Studies with APP mutated close to the α-secretase cleavage site suggested that local α-helicity contributes to cleavage efficiency presumably by direct interaction of the endoprotease to this structure (3).
Recently, evidence was provided that TACE, also known as ADAM 17, may be involved in regulated α-secretase cleavage of APP (46). Disruption of the TACE gene abolished the augmented secretion of APPs in mouse fibroblasts in response to phorbol ester. However, basal formation and secretion of APPs was unaffected in cells derived from knock-out mice. Furthermore, there was a different effect of three hydroxamic acid-based inhibitors toward the TNF-α converting enzyme compared with α-secretase (47). Thus, TACE would appear to be distinct from the α-secretase in respect to basal activity and inhibitor spectrum but shares the phorbol ester-enhanced secretase activity with ADAM 10. Although human TACE and human ADAM 10 have only 21% amino acid identity, there is evidence that their catalytic domains have some common structural properties (48).
Two yeast aspartyl proteases have recently been described which cleave APP in yeast at the expected α-secretase cleavage site (49). Thus, with the observation of heterogeneity of cleavages in this site (14, 15), this could indicate that apart from ADAM 10 other enzymes may act as α-secretases.
The identification of ADAM 10 as an enzyme with constitutive and regulated α-secretase activity provides a target for increasing its expression and activity, which might prove beneficial in Alzheimer’s disease.
Acknowledgments
We thank Dr. S. Rose-John (University Mainz) for the peptide with IL-6 receptor sequence, Dr. T. A. Link (University Frankfurt/M) for his advice concerning CD measurements, Dr. H. Steiner (University of Heidelberg) for his help in immunoprecipitation experiments, and British Biotech for supplying the inhibitor BB-3103. We also appreciate the technical help and computing advice of F. Bender (University Mainz). This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 1571), by the Fonds der Chemischen Industrie, and by the Naturwissenschaftlich-Medizinisches Forschungszentrum (University Mainz). S.L. was supported by the Graduiertenkolleg, “Proteinstrukturen, Dynamik und Funktion” (University Frankfurt/M).
ABBREVIATIONS
- Aβ
amyloid β peptide
- ADAM
a disintegrin and metalloprotease
- APP
amyloid precursor protein
- APPsα
α-secretase cleaved soluble APP
- HA
hemagglutinin
- HEK
human embryonic kidney
- PMA
phorbol-12-myristate 13-acetate
- TACE
tumor necrosis factor α converting enzyme
- TNF-α
tumor necrosis factor α
- PVDF
poly(vinylidene difluoride)
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Haass C, Selkoe D J. Cell. 1993;75:1039–1042. doi: 10.1016/0092-8674(93)90312-e. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D J. J Biol Chem. 1996;271:18295–18298. doi: 10.1074/jbc.271.31.18295. [DOI] [PubMed] [Google Scholar]
- 3.Sisodia S S. Proc Natl Acad Sci USA. 1992;89:6075–6079. doi: 10.1073/pnas.89.13.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haass C, Koo E H, Mellon A, Hung A Y, Selkoe D J. Nature (London) 1992;357:500–503. doi: 10.1038/357500a0. [DOI] [PubMed] [Google Scholar]
- 5.Ikezu T, Trapp B D, Song K S, Schlegel A, Lisanti M P, Okamoto T. J Biol Chem. 1998;273:10485–10495. doi: 10.1074/jbc.273.17.10485. [DOI] [PubMed] [Google Scholar]
- 6.Kuentzel S L, Ali S M, Altman R A, Greenberg B D, Raub T J. Biochem J. 1993;295:367–378. doi: 10.1042/bj2950367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Strooper B, Umans L, Van Leuven F, Van Den Berghe H. J Cell Biol. 1993;121:295–304. doi: 10.1083/jcb.121.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sambamurti K, Shioi J, Anderson J P, Pappolla M A, Robakis N K. J Neurosci Res. 1992;33:319–329. doi: 10.1002/jnr.490330216. [DOI] [PubMed] [Google Scholar]
- 9.Tomita S, Yutaka K, Suzuki T. J Biol Chem. 1998;273:6277–6284. doi: 10.1074/jbc.273.11.6277. [DOI] [PubMed] [Google Scholar]
- 10.Weidemann A, König G, Bunke D, Fischer P, Salbaum J M, Masters C L, Beyreuther K. Cell. 1989;57:115–126. doi: 10.1016/0092-8674(89)90177-3. [DOI] [PubMed] [Google Scholar]
- 11.Haass C, Hung A Y, Schlossmacher M G, Teplow D B, Selkoe D J. J Biol Chem. 1993;268:3021–3024. [PubMed] [Google Scholar]
- 12.Esch F S, Keim P S, Beattie E C, Blacher R W, Culwell A R, Oltersdorf T, McClure D, Ward P J. Science. 1990;248:1122–1124. doi: 10.1126/science.2111583. [DOI] [PubMed] [Google Scholar]
- 13.Pasternack J M, Palmert M R, Usiak M, Wang R, Zürcher-Neely H, Gonzalez-DeWhitt P A, Fairbanks M B, Cheung T, Blades D, Heinrikson R L, et al. Biochemistry. 1992;31:10936–10940. doi: 10.1021/bi00159a038. [DOI] [PubMed] [Google Scholar]
- 14.Zhong Z, Higaki J, Murakami K, Wang Y, Catalano R, Quon D, Cordell B. J Biol Chem. 1994;269:627–632. [PubMed] [Google Scholar]
- 15.Simons M, De Strooper B, Multhaup G, Tienari P J, Dotti C G, Beyreuther K. J Neurosci. 1996;16:899–908. doi: 10.1523/JNEUROSCI.16-03-00899.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black R A, Rauch C T, Kozlosky C J, Peschon J J, Slack J L, Wolfson M F, Castner B J, Stocking K L, Reddy P, Srinivasan S, et al. Nature (London) 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 17.Moss M L, Jin S-L C, Milla M E, Bickett D M, Burkhart W, Carter H L, Chen W-J, Clay W C, Didsbury J R, Hassler D, et al. Nature (London) 1997;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 18.Blobel C P, Wolfsberg T G, Turck C W, Myles D G, Primakoff P, White J M. Nature (London) 1992;356:248–252. doi: 10.1038/356248a0. [DOI] [PubMed] [Google Scholar]
- 19.Yagami-Hiromasa T, Sato T, Kurisaki T, Kamijo K, Nabeshima Y-I, Fujisawa-Sehara A. Nature (London) 1995;377:652–656. doi: 10.1038/377652a0. [DOI] [PubMed] [Google Scholar]
- 20.Rooke J, Pan D, Xu T, Rubin G M. Science. 1996;273:1227–1231. doi: 10.1126/science.273.5279.1227. [DOI] [PubMed] [Google Scholar]
- 21.Pan D, Rubin G M. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 22.Qi H, Rand M D, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- 23.Chantry A, Gregson N A, Glynn P. J Biol Chem. 1989;264:21603–21607. [PubMed] [Google Scholar]
- 24.Howard L, Lu X, Mitchell S, Griffiths S, Glynn P. Biochem J. 1996;317:45–50. doi: 10.1042/bj3170045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts S B, Ripellino J A, Ingalls K M, Robakis N K, Felsenstein K M. J Biol Chem. 1994;269:3111–3116. [PubMed] [Google Scholar]
- 26.Howard L, Glynn P. Methods Enzymol. 1995;248:388–395. doi: 10.1016/0076-6879(95)48025-0. [DOI] [PubMed] [Google Scholar]
- 27.Wessel D, Flügge U I. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 28.Haass C, Hung A Y, Selkoe D J. J Neurosci. 1991;11:3783–3793. doi: 10.1523/JNEUROSCI.11-12-03783.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haass C, Schlossmacher M G, Hung A Y, Vigo-Pelfrey C, Mellon A, Ostaszewski B L, Lieberburg I, Koo E H, Schenk D, Teplow D B, et al. Nature (London) 1992;359:322–325. doi: 10.1038/359322a0. [DOI] [PubMed] [Google Scholar]
- 30.Kirsch J, Betz H. J Neurosci. 1995;15:4148–4156. doi: 10.1523/JNEUROSCI.15-06-04148.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendricks L, Duijn C, Cras P, Cruts M, van Hul W, van Harskamp F, Warren A, McInnis M, Antonarakis S, Martin J-J, et al. Nat Genet. 1992;1:218–221. doi: 10.1038/ng0692-218. [DOI] [PubMed] [Google Scholar]
- 32.Haass C, Hung A Y, Selkoe D J, Teplow D B. J Biol Chem. 1994;269:17741–17748. [PubMed] [Google Scholar]
- 33.Capell A, Teplow D B, Citron M, Selkoe D J, Haass C. Amyloid. 1996;3:150–155. [Google Scholar]
- 34.Ehlers M R W, Schwager S L U, Scholle R R, Manji G A, Brandt W F, Riordan J F. Biochemistry. 1996;35:9549–9559. doi: 10.1021/bi9602425. [DOI] [PubMed] [Google Scholar]
- 35.Müllberg J, Oberthür W, Lottspeich F, Mehl E, Dittrich E, Graeve L, Heinrich P C, Rose-John S. J Immunol. 1994;152:4958–4968. [PubMed] [Google Scholar]
- 36.Lunn C A, Fan X, Dalie B, Miller K, Zavodny P J, Narula S K, Lundell D. FEBS Lett. 1997;400:333–335. doi: 10.1016/s0014-5793(96)01410-x. [DOI] [PubMed] [Google Scholar]
- 37.Rosendahl M S, Ko S C, Long D L, Brewer M T, Rosenzweig B, Hedl E, Anderson L, Pyle S M, Moreland J, Meyers M A, et al. J Biol Chem. 1997;272:24588–24593. doi: 10.1074/jbc.272.39.24588. [DOI] [PubMed] [Google Scholar]
- 38.Middelhoven P J, Ager A, Roos D, Verhoeven A J. FEBS Lett. 1997;414:14–18. doi: 10.1016/s0014-5793(97)00959-9. [DOI] [PubMed] [Google Scholar]
- 39.Lum L, Reid M S, Blobel C P. J Biol Chem. 1998;273:26236–26247. doi: 10.1074/jbc.273.40.26236. [DOI] [PubMed] [Google Scholar]
- 40.Parvathy S, Hussain I, Karran E H, Turner A, Hooper N M. Biochemistry. 1998;37:1680–1685. doi: 10.1021/bi972034y. [DOI] [PubMed] [Google Scholar]
- 41.Hung A Y, Haass C, Nitsch R M, Qin W Q, Citron M, Wurtman R J, Growdon J H, Selkoe D J. J Biol Chem. 1993;268:22959–22962. [PubMed] [Google Scholar]
- 42.Buxbaum J D, Koo E H, Greengard P. Proc Natl Acad Sci USA. 1993;90:9195–9198. doi: 10.1073/pnas.90.19.9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Strooper B, Simons M, Multhaup G, Van Leuven F, Beyreuther K, Dotti C D. EMBO J. 1995;14:4932–4938. doi: 10.1002/j.1460-2075.1995.tb00176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LeBlanc A C, Xue R, Gambetti P. J Neurosci. 1996;66:2300–2310. doi: 10.1046/j.1471-4159.1996.66062300.x. [DOI] [PubMed] [Google Scholar]
- 45.Chapman R E, Munro S. EMBO J. 1994;13:2305–2312. doi: 10.1002/j.1460-2075.1994.tb06514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buxbaum J D, Liu K-N, Luo Y, Slack J L, Stocking K L, Peschon J J, Johnson R S, Castner B J, Cerretti D P, Black R A. J Biol Chem. 1998;273:27765–27767. doi: 10.1074/jbc.273.43.27765. [DOI] [PubMed] [Google Scholar]
- 47.Parvathy S, Karran E H, Turner A J, Hooper N M. FEBS Lett. 1998;431:63–65. doi: 10.1016/s0014-5793(98)00726-1. [DOI] [PubMed] [Google Scholar]
- 48.Maskos K, Fernandez-Catalan C, Huber R, Bourenkov G P, Bartunik H, Ellestad G A, Reddy P, Wolfson M F, Rauch C T, Castner B J, et al. Proc Natl Acad Sci USA. 1997;95:3408–3412. doi: 10.1073/pnas.95.7.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komano H, Seeger M, Gandy S, Wang G T, Krafft G A, Fuller R S. J Biol Chem. 1998;273:31648–31651. doi: 10.1074/jbc.273.48.31648. [DOI] [PubMed] [Google Scholar]