Abstract
Alterations of human chromosome 8p occur frequently in many tumors. We identified a 1.5-Mb common region of allelic loss on 8p22 by allelotype analysis. cDNA selection allowed isolation of several genes, including FEZ1. The predicted Fez1 protein contained a leucine-zipper region with similarity to the DNA-binding domain of the cAMP-responsive activating-transcription factor 5. RNA blot analysis revealed that FEZ1 gene expression was undetectable in more than 60% of epithelial tumors. Mutations were found in primary esophageal cancers and in a prostate cancer cell line. Transcript analysis from several FEZ1-expressing tumors revealed truncated mRNAs, including a frameshift. Alteration and inactivation of the FEZ1 gene may play a role in various human tumors.
Frequent loss of heterozygosity (LOH) at specific chromosomal regions in certain tumors implies the presence of suppressor genes (1–5). Recent allelotyping studies have shown that allelic losses on the short arm of chromosome 8, particularly at bands 21–22, frequently are associated with various tumors, including prostate cancer (6, 7), breast cancer (8), head and neck squamous cell carcinoma (9–11), urinary bladder carcinoma (12), hepatocellular carcinoma (13), and hematological malignancies (14). A recent genome-wide search for LOH in breast cancer showed that 8p is one of the most frequently altered chromosome regions (15); LOH at 8p21–22 was associated with the invasive behavior of breast cancer (16, 17). Loss at 8p21.3–22 has been shown to be associated strongly with prostate cancer progression (18). These observations suggest that chromosome region 8p21–22 plays an important role in the development of various tumors, including prostate and breast cancers.
Functional evidence of the presence of tumor suppressor gene(s) at 8p was shown experimentally by chromosome transfer into tumor cells (19). Somatic cell hybrids of cancer cells with transferred normal human chromosome 8p22–23 fragments lost their ability to produce tumors (19). Additional microcell fusion experiments suggested the possible location of metastasis suppressor gene(s) at 8p (20–22). It is possible that two or more genes at 8p may be involved in suppression of cancer development.
Efforts toward positional cloning of the suppressor gene(s) allowed the isolation of a candidate tumor suppressor gene, N33, at 8p22 near the macrophage-scavenger-receptor (MSR) gene locus (23–26). The N33 gene was silenced in several cancer cells, although no point mutations were found. Another candidate gene, PRLTS, at 8p21.3–22 showed point mutations in four cancer cases (27, 28). The frequency of alterations in this gene was, however, very low (28). Alterations of N-acetyltransferase (NAT)1 and NAT2 genes at 8p22 have also been studied in cancer, because of their carcinogen-metabolizing action (29, 30). The results showed no abnormality in cancer cells (29, 30).
Esophageal cancer occurs worldwide, and its incidence is increasing in the Western world (31–33). We performed allelotyping to identify a common region of loss at 8p in primary esophageal squamous cell carcinomas. We identified a common region of LOH at 8p22 around marker D8S261, which overlaps with the target region in other tumors, including prostate and breast cancer (6–9, 15). This region is >2 Mb centromeric to the MSR region (23–26). To clone the genes present in this region, cDNA selection, CpG island cloning, and shotgun sequencing were carried out. Analysis of cDNAs in human tumors and tumor-derived cell lines indicates that we cloned a gene, FEZ1, which is altered in many tumors, including esophageal, prostate, and breast cancer.
MATERIALS AND METHODS
Cell Lines and Tissues.
Esophageal cancer cell lines were cultured in RPMI 1640 with 10% FBS (34). The other cancer cell lines were obtained from the American Type Culture Collection and maintained. Seventy two primary esophageal cancer samples and flanking normal portions, as well as 39 breast, 24 prostate, and 8 ovarian cancers, were obtained surgically from patients with their informed consent.
LOH Study.
PCR amplifications using 5′-fluorescein phosphoramidite- or 5′-tetrachloro-fluorescein phosphoramidite-labeled primers for microsatellite loci (Research Genetics, Huntsville, AL) with tumor and normal template DNAs were performed as reported (35), with minor modifications. Briefly, PCRs were done in a 20 μl of buffer containing 2.5 mM MgCl2, 1.5 mM dNTP, and 0.5 unit of Ampli Taq Gold (Perkin–Elmer). PCR conditions were as follows; after 95°C for 12 min, a total of 30 cycles consisting of 10 cycles at 94°C for 15 sec, 55–58°C annealing for 15 sec, and 72°C for 30 sec, and 20 cycles at 89°C for 15 sec, 55–58°C annealing for 15 sec, and 72°C for 30 sec, followed by 72°C for 30 min. After heat denaturation, the samples were loaded on a 6% denaturing gel on the Applied Biosystems 373 DNA sequencer. Data collection and fragment analysis were done with the Applied Biosystems Prism Genescan and the Applied Biosystems prism genotyper analysis software (Perkin–Elmer/Applied Biosystems). Cases were judged as LOH when an allele peak signal from tumor DNA was reduced by 50% compared with the normal counterpart. When tumors showed 40–60% reduction of a normal counterpart, the analyses were repeated two more times, and average reductions were used as final data.
Yeast Artificial Chromosome (YAC) and Bacterial Artificial Chromosome (BAC) DNAs.
PCR amplifications were done to screen human YAC and BAC libraries (Research Genetics). YAC clones were embedded into agarose and separated by pulse-field gel electrophoresis (36, 37). After the gel electrophoresis, the YAC DNAs were cut out from the gels. BAC DNAs were purified by using the Qiagen (Chatsworth, CA) purification kit. BAC DNAs were sequenced with T7 and SP6 primers. Southern blot hybridization and PCR analysis resulted in overlapping clones. From these results, contigs were constructed.
cDNA Selection.
Gels containing YAC DNAs were digested with MboI and were extracted from the gel with Gene Clean III (Bio 101). Two deoxyoligonucleotides, 5′-GATCTCGACGAATTCGTGAGACCT-3′ and 5′-TGGTCTCACGAATTCGTCGA-3′, were annealed to make an adapter-linker, which was ligated to the digested genomic DNA. PCR amplifications with 15 cycles were performed with 5′-biotinylated primer corresponding with the adapter-linker. cDNAs were reverse-transcribed from prostate poly(A)+ RNA (CLONTECH) with NotI-primer adaptor/oligo(dT)-primers (Superscript Plasmid system; GIBCO/BRL). After the first strand cDNA end was blunted, a SalI adaptor (GIBCO/BRL) was ligated to cDNAs. cDNAs were amplified by PCR with the SalI adapter primers, and the products were cleaned with the Qiagen PCR purification column.
The methods of blocking, hybridization, and washing (26, 38) were adapted with minor modifications. Repetitive sequences were blocked with equal amounts (wt/wt) of Cot-1 DNA (GIBCO/BRL). Biotin-labeled genomic DNAs and blocked cDNA were adjusted to 120 mM NaPO4 (pH 7) and 1 mM EDTA (pH 8) and a DNA concentration (excluding Cot-1) of approximately 160 μg/ml. Reactions were incubated at 60°C for 60 h (Cot = 120). For washing, 10 μl of avidin-coated magnetic bead suspension (Dynabeads M-280; Dynal, Great Neck, NY), which was premixed with sonicated salmon sperm DNA, was incubated with complete hybridization reactions in TE (Tris-EDTA) buffer and 1M NaCl at room temperature for 30 min. Two washes were performed with 0.1× standard saline citrate (1× = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7) and 0.1% SDS for 15 min at room temperature, and three washes were done at 65°C. Bound cDNAs were eluted from beads, neutralized, and cleaned with the Qiagen PCR purification column. The cDNAs were reamplified by PCR in the same conditions as the first round, and the amplified cDNAs were digested with SalI and NotI and were cloned directly into pSPORT1 vector (GIBCO/BRL).
Cloning of CpG Island and Shotgun Sequencing of BAC.
BAC DNAs were digested with BssHII or SacII, and with Sau3AI (39). The products were ligated into pBK-CMV (Stratagene), predigested with BssHII or SacII and BamHI. The products were subcloned and sequenced. Shotgun sequencing was performed as described (40). Clones from the shotgun libraries were sequenced to identify candidate cDNA sequences.
Reverse Transcription (RT)–PCR, Rapid Amplification of cDNA Ends, and cDNA Library Screening.
cDNA was synthesized from 2 μg of total RNA or 150 ng of poly(A)+ RNA with Superscript II (GIBCO/BRL) at 42°C or 50°C and subjected to PCR amplifications with specific primers. The rapid amplification of cDNA ends was performed using cDNAs from human testes and brain (CLONTECH). The cDNA libraries from human testes and brain (CLONTECH) were screened.
Nucleotide Sequence.
Nucleotide sequence information was obtained by DNA sequencing. Eleven pairs of primers for FEZ1 exons 1–3 were used for PCR with genomic DNA. The PCRs were done with the same conditions as those described for the LOH studies, except that 4% (wt/wt) dimethyl sulfoxide was added, and PCR amplifications were a total of 35 cycles. The PCR products were cleaned with the Qiagen PCR purification column and sequenced. Sequencing reactions and analysis were performed by using the Applied Biosystems Prism BigDye terminator reaction chemistry on a Perkin–Elmer Gene Amp PCR system 9600 and the Applied Biosystems Prism 377 DNA sequencing system.
RESULTS AND DISCUSSION
Deletion Analysis of Chromosome 8p in Cancer.
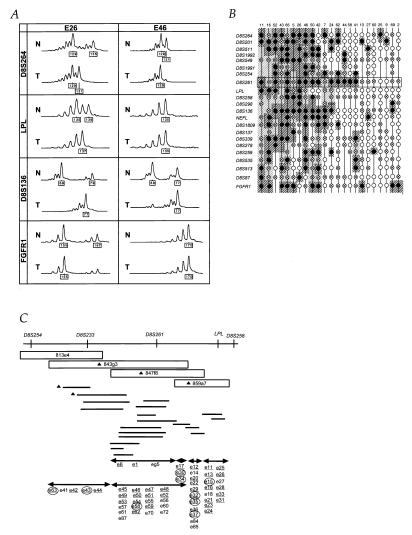
DNAs from 53 primary esophageal cancers and matched normal tissues were analyzed for allele loss at 22 microsatellite loci on chromosome 8p. Representative data are shown in Fig. 1A. Allelic loss was assessed by the reduction of the signal intensity of an allele, compared with its normal counterpart (35). The variability of relative intensity of tumor alleles is caused by either intratumoral heterogeneity or normal tissue contamination. Histological examination showed that different tumors had different ratios of neoplastic to stromal cells and to infiltrating lymphocytes. The apparent incomplete allele loss seen in some samples, such as E46 at D8S136 and D8S264 (Fig. 1A) likely is caused by the contamination of normal cells. The allelic losses for each matched DNA pair are summarized in Fig. 1B. Twenty-three patients (43%) showed loss of an allele at one or more loci on 8p. Sixteen of 23 tumors (70%) showed a commonly lost 1.5-Mb region near the D8S261 loci, and 14 patients (61%) had potential common LOH regions near D8S254 (Fig. 1B). These data suggest that two tumor suppressor genes are located in the chromosome 8p22–23 region. We focused on the more frequently affected 8p22 region around D8S261.
Figure 1.
LOH at human chromosome 8p in primary esophageal cancer samples and genomic contigs at 8p22. (A) Representative LOH analysis from two cases, E26 and E46. Fluorescent PCR products from normal (N) and tumor (T) DNAs were analyzed. The x axis shows DNA fragment size in bp (depicted in boxes, compared with size markers). Patient E26 was informative for all four markers; LOH for D8S264, LPL, and D8S136 and allelic retention in FGFR1 (because a 167-bp peak in tumor is more than 50% of that in normal). Patient E46 was informative at D8S264 and D8S136, and one allele at each was lost, whereas LPL and FGFR1 loci were homozygous. (B) Summary of LOH analyses. All the patients with loss at least at one locus are shown. The closed circles indicate loss of an allele, the circles with crosses indicate noninformation because of homozygosity, and the open circles depict the retention of both alleles. The dark hatched areas indicate regions of allele loss, whereas the light hatched areas show regions of noninformation within allele loss areas. Further studies focused on the region near the marker D8S261 locus shown in a boxed area. The column numbers correspond to patients, whereas the row numbers indicate polymorphic markers. (C) Genomic contigs at 8p22. The upper line depicts the location of polymorphic loci on 8p. YAC (boxes) and BAC (horizontal lines) contigs were constructed. The cDNA selection was performed on three YAC DNAs (triangles). The shotgun sequencing were carried out on BACs (triangles). Eighty-seven potentially expressed sequences were isolated and located within the contigs, which are shown in the lower portion. The underlined characters are the sequences that are expressed in normal tissues. A candidate fragment, e37, corresponds to the F37 cDNA described in text. Areas with arrows show locations of candidate cDNAs. After expression analysis in tumor and normal tissues, nine cDNAs (circled characters) were subjected to partial cloning and further analysis.
YAC and BAC Contigs.
YAC clones were isolated, and a genomic YAC contig of the region around the D8S261 marker was constructed (Fig. 1C). To analyze the commonly lost region, we isolated BAC clones, and a BAC contig, including markers D8S233 and D8S261, was constructed. The overlaps of BAC clones were confirmed by Southern blot or by PCR amplification.
Candidate cDNAs Isolation and Mapping.
We used three approaches to isolate candidate cDNAs from the target region: CpG island cloning, shotgun sequencing, and cDNA selection (Fig. 1C). Two candidates for CpG islands were identified from the region near marker D8S233 by CpG island cloning. The BACs were partially sequenced by the shotgun method to look for sequences matching expressed sequence tags in the database. These approaches resulted in identifying two expressed sequence tags from this region. We also performed cDNA selection for three YAC templates, which covered D8S261 and LPL. We picked 400 clones per YAC from the cDNA selected libraries and sequenced all the clones using two vector primers. Fifty percent of clones were ribosomal or mitochondria-related genes, and the remainders were classified and analyzed. A total of 123 clones of 400–800 bp were selected.
Eighty-seven potentially expressed clones were mapped in the YAC contig (Fig. 1C). To choose clones that are expressed, we performed RT-PCR with RNAs from normal adult tissues, including prostate. RT-PCR analysis revealed that 43 clones were expressed in normal tissues. To select clones that showed reduced expression in tumor cells, RT-PCR amplifications were performed with cDNA from prostate cancer cell lines. Nine clones showed absence or reduction of expression in cancer cells. The rapid amplification of cDNA ends method was carried out, and the sequences of six clones were extended successfully. Northern blot analyses were performed with these cDNA probes. F30 and F34 clones showed similar expression patterns, including a major transcript of about 10 kb and a smaller transcript of 2 kb in testes, suggesting that these two cDNAs were from the same gene. Alterations of expression of F30/F34 and three other clones were not remarkable in cancer (data not shown). On the other hand, Northern blot analysis using the F37 clone, obtained by hybrid selection, showed a 6.8-kb transcript in normal epithelial tissues, the expression of which could not be detected in the LNCaP prostate cancer cell line.
Cloning of the Full-Length F37 Gene.
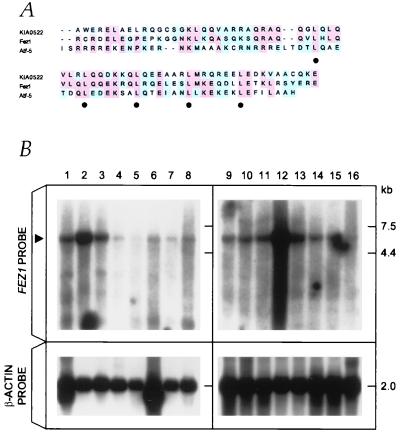
A human testes cDNA library of 6 × 106 clones was screened with the F37 probe, and the 5′ end was obtained by the rapid amplification of cDNA ends procedures. The accuracy of the cDNA sequence was confirmed by RT-PCR, sequencing, and Northern blot analysis using several different probes from separate regions of the F37 cDNA. Exon/intron boundaries were identified by sequencing the corresponding BAC clones. The chromosomal location of the F37 gene was confirmed by the presence of the F37 gene fragment in a radiation hybrid panel (Gene Bridge 4 Human/Hamster RH Panel, Research Genetics). The result showed that the F37 gene is located within 3.36 centiRays from WI-5962 at chromosome 8p22. The F37 cDNA is approximately 6.8 kb, including a 1,788-bp ORF, which encoded a 596-aa protein of 67 kDa. Homology search of databases showed that the F37 sequence contains a leucine-zipper motif and it has 32% identity to the DNA-binding domain of a cAMP-responsive activating-transcription factor, Atf-5 (Fig. 2A). The homology search also showed that the F37 protein has 38% identity to the KIAA0552 protein, which consists of 673 aa. The motif analysis (Searching Protein and Nucleic Acid Sequence Motifs in Genome Net) showed a predicted cAMP-dependent phosphorylation site, Ser-29, and a predicted tyrosine kinase phosphorylation site, Tyr-67. The ORF was composed of coding exons 1–3 (Fig. 3C). In vitro transcription/translation experiments showed that the F37 gene can be transcribed and translated into a protein (data not shown). The F37 gene was designated FEZ1 (F37/Esophageal cancer-related gene-coding leucine-zipper motif).
Figure 2.
The predicted FEZ1 amino acid sequence and the expression. (A) Comparison of the predicted amino acid sequences corresponding to the DNA-binding and leucine-zipper regions. Comparison of Fez1 (amino acids 301–369 are depicted) with Atf-5 and KIAA0522 proteins are shown. Identical (red shading) or similar (blue shading) residues among Fez1 and the other two proteins are indicated. Gaps introduced by the fasta program are represented by spaces. Closed circles denote repeated leucine residues. (B) FEZ1 gene expression. Northern blot analysis of normal tissue RNAs. Probes used were the FEZ1 ORF probe (upper) and the control β-actin probe (lower). Poly(A)+ RNA (5 μg) was loaded. The arrowhead on the left indicates the 6.8-kb FEZ1 transcript. RNAs in lanes were from: 1, heart; 2, brain; 3, placenta; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; 8, pancreas; 9, spleen, 10, thymus; 11, prostate; 12, testes; 13, ovary; 14, small intestine; 15, colon; 16, peripheral blood lymphocyte.
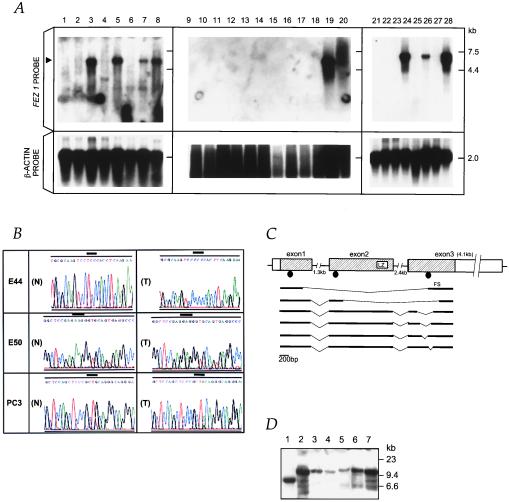
Figure 3.
Alterations of FEZ1 in tumors. (A) FEZ1 gene expression in cancers. Probes used were a FEZ1 cDNA probe (Upper) and a control β-actin probe (Lower). Poly(A)+ RNA (5 μg) from cancer cell lines was loaded. The arrowhead on the left indicates the 6.8-kb transcript of FEZ1. RNAs were from: esophageal cancer cell lines KYSE170 (lane 1), TE12 (lane 2), TE8 (lane 3), and TE3 (lane 4); prostate cancer cell lines DU145 (lane 5), LNCaP (lane 6), PC3 (lane 7), and normal prostate (lane 8); breast cancer cell lines MB231 (lane 9), SKBr3 (lane 10), BT549 (lane 11), HBL100 (lane 12), MB436 (lane 13), BT20 (lane 14), MB543 (lane 15), MB175 (lane 16), MCF7 (lane 17), and T47B (lane 18); normal breast (lane 19); and total RNA of normal breast (lane 20); promyelocytic leukemia cell line HL60 (lane 21); cervical cancer cell line HeLa S3 (lane 22); chronic myelogenous leukemia cell line K562 (lane 23); lymphoblastic leukemia cell line MOLT4 (lane 24); Burkitt’s lymphoma cell line Raji (lane 25); colorectal adenocarcinoma cell line SW480 (lane 26); lung cancer cell line A549 (lane 27); and melanoma cell line G361 (lane 28). (B) Sequence chromatograms of three mutations. A point mutation (TCC/Ser → CCC/Pro) at codon 29 was found (T) in an primary esophageal cancer, E44. As controls, normal sequences from normal DNA from patient E44 and BAC sequence (N) were analyzed, both producing the same result. A bold line indicates the altered codon. In primary esophageal cancer, E50, a point mutation (AAG/Lys → GAG/Glu) at codon 119 was found (T). The normal BAC sequence is shown in (N). The point mutation (CAG/Gln → TAG/STOP) at codon 501 was observed in prostate cancer PC3 cells (T). Note that the sequence chromatogram from this cell line is shown in the 3′ to 5′ direction. Repeated sequencing showed a weak signal corresponding to guanine (G) within a large adenine (A) signal in the first nucleotide at codon 501, suggesting the retention of the normal allele in a fraction of the cancer cells. Expression analysis with RT-PCR detected only the mutated transcript in this cell line. The normal BAC sequence is shown in (N). (C) Truncated FEZ1 transcripts observed in cancers. The normal exon/intron structure (top drawing) was determined by sequencing of brain, prostate, and esophagus cDNAs and by sequencing FEZ1 gene in BAC. The boxes indicate exons, and the shaded areas show the ORF of 1,788 bp. The introns are depicted by horizontal lines. Aberrant transcripts observed in tumors are depicted in bold lines. The location of mutations are shown as closed circles. LZ, leucine-zipper motif; FS, frameshift. (D) Southern blot analysis of the FEZ1 gene locus. High-Mr DNAs from cancer cells were cleaved with EcoRI, separated electrophoretically, transferred to a nylon membrane, and probed with the 1.7-kb FEZ1 ORF probe. DNAs (10 μg/lane) were from lanes: 1, MB436S; 2, normal; 3, MB231; 4, MB361; 5, TE8; 6, TE3; 7, normal (different origin from normal DNA in lane 2).
Expression of the FEZ1 Gene in Normal and Tumor Tissues.
Northern blot analysis revealed that the FEZ1 gene is expressed almost ubiquitously in normal tissues, although expression in testes was most abundant (Fig. 2B). We also analyzed the expression of the FEZ1 gene in human tumors, including 41 cancer-derived cell lines and 25 primary tumors, by Northern blotting and by RT-PCR amplifications (Fig. 3A; summarized in Table 1). The results showed that FEZ1 expression was undetectable in 31 cancer cell lines (76%) and 16 primary tumors (64%). FEZ1 expression was not detected in any of the breast cancer cell lines (15 of 15) or primary tumors (10 of 10) studied. To exclude the possibility of involvement of some alternative splicing variants, which may affect the expression analysis, we performed Northern blot analysis with three different probes (from the ORF, from the 3′-noncoding sequence just downstream of the ORF, and from the 3′-noncoding terminal sequence). The results showed no differences in detection of transcripts by three probes, suggesting that the FEZ1 expression was absent in the cell lines and tumors examined. To exclude the possibility that normal stromal cells, but not epithelial cells, may express FEZ1, we analyzed the FEZ1 expression in normal breast epithelial cells and fibroblasts and in normal prostate epithelial cells (Clonetics, San Diego). The results with RT-PCR amplifications showed that FEZ1 was expressed in all of these normal cells, although no expression was observed in breast and prostate cancer cells (data not shown).
Table 1.
Summary of FEZ1 gene expression in various cancers
| Origin of cancer samples | No. of cases analyzed | Cases expressing FEZ1 mRNAs | No. of cases with aberrant size transcripts (case names) |
|---|---|---|---|
| Esophagus | |||
| Cell lines | 4 | 1 | 1 (TE8) |
| Primary tumors | 12 | 9* | 4 (E16, E26, E41, E62) |
| Gastric cell lines | 8 | 3* | Not done |
| Colon cell lines | 3 | 2 | 1 (SW480) |
| Prostate | |||
| Cell lines | 3 | 2 | 1 (DU145) |
| Primary tumors | 3 | 0* | |
| Breast | |||
| Cell lines | 15 | 0 | |
| Primary tumors | 10 | 0* | |
| Hematopoietic cell lines | 5 | 1 | 1 (MOLT4) |
| Lung cell lines | 1 | 0 | |
| Melanoma cell lines | 1 | 1 | 1 (G361) |
| Cervical cell lines | 1 | 0 |
The FEZ1 expression was analyzed by Northern blot or RT-PCR.
, Analyzed by RT-PCR.
Mutations of the FEZ1 Gene in Tumors.
The nucleotide sequence of the FEZ1 gene ORF (exons 1–3) was analyzed in a total of 194 cancers, from 72 primary esophageal cancers, 18 esophageal cancer cell lines, 24 primary prostate cancers, 3 prostate cancer cell lines, 39 primary breast cancers, 25 breast cancer cell lines, 8 primary ovarian cancers, 4 leukemic cell lines, and 1 cervical cancer cell line, regardless of the presence or absence of FEZ1 expression. DNA sequencing was performed directly with the purified PCR products, not the subcloned PCR products. The data were confirmed by sequencing of duplicate PCR amplification products and by sequencing the antisense strands with reverse primers. For primary tumor cases with mutations, normal DNA from the same patient was also analyzed. We found three point mutations in two primary esophageal cancers and in a prostate cancer cell line (Fig. 3 B and C). In a primary esophageal tumor, E44, alteration of TCC to CCC at codon 29 resulted in the substitution of Ser-29 with Pro-29, which is a predicted cAMP-dependent kinase phosphorylation site, whereas the sequences from both the normal DNA of the same patient and human BAC clones showed no alterations. In another primary esophageal cancer, E50, alteration of AAG/Lys to GAG/Glu at codon 119 was found. Our LOH study indicated that these two patients had allelic losses at the D8S261 marker. Thus, these tumor cells retained the mutated FEZ1 allele and lost the normal FEZ1 allele. The third point mutation was the change of CAG/Gln to TAG/Stop at codon 501 in a prostate cancer cell line, PC3, which resulted in coding of a putative 166-aa protein lacking the C terminus.
Aberrant Transcripts of FEZ1 Gene from Tumors.
We analyzed by RT-PCR the cDNA sequences in the ORF from a fraction of the tumors that expressed the FEZ1 gene (Fig. 3C; Table 2). In esophageal cancer, prostate cancer, melanoma, and hematological malignancies, several internally truncated transcripts were identified. Sequences from normal brain and prostate, as well as normal esophagus from seven individuals and from matched normal cDNA of patients, E16, E26, and E41, showed no alterations, except that one of 12 clones from testes cDNA showed a deleted sequence (nucleotides 1,441–1,527 in the ORF were deleted). The transcript from two independent esophageal cancers showed a frameshift within the ORF, which resulted in coding a short 76-aa protein (Fig. 3C). We analyzed the nucleotide sequences around “break points” of the deleted cDNAs and compared them to sequences from the full-length cDNA. The results showed that all of the acceptor sites contain the intronic AG flanking the exons, and these are almost compatible with the conserved sequences frequently observed in the eukaryote exon/intron boundary structure (41, 42), suggesting that the deleted transcripts in several tumors may be produced by splicing. We also analyzed the allelic expression status of the FEZ1 gene with a polymorphic site in the 3′-noncoding cDNA region (the 2,134th nucleotide from the first codon). The results showed that, in four informative, normal primary tissues analyzed, the FEZ1 gene was transcribed from both alleles, i.e., not imprinted, whereas the expression in the FEZ1-expressing cancer cells was from a single allele, probably because of the allelic losses (data not shown).
Table 2.
FEZ1 aberrant transcripts observed in cancers
| Tumor | Deletion | Results | Affected exons | Putative protein coded in-frame |
|---|---|---|---|---|
| E16 | 156-1542 | FS | Ex1,2,3 | Zip(−) |
| E26 | 558-1715 | IF | Ex2,3 | Zip(−) |
| E41 | 558-1715 | IF | Ex2,3 | Zip(−) |
| E62 | 558-1715 | IF | Ex2,3 | Zip(−) |
| TE8 | 156-1542 | FS | Ex1,2,3 | Zip(−) |
| 1402-1578 | IF | Ex3 | Zip(+) | |
| DU145 | 1366-1641 | IF | Ex3 | Zip(+) |
| 1402-1578 | IF | Ex3 | Zip(+) | |
| MOLT4 | 1402-1578 | IF | Ex3 | Zip(+) |
| G361 | 1417-1515 | IF | Ex3 | Zip(+) |
| 1516-1584 | IF | Ex3 | Zip(+) |
The positions of the first and last nucleotides of deletions are shown according to the nucleotide number counted from the first coding codon. IF, in-frame; FS, frameshift; Zip(+), putative Fez1 protein coded in-frame with leucine zipper: Zip(−), protein without leucine zipper.
Genomic Analysis of the FEZ1 Gene Region.
Several tumor suppressor genes are associated with frequent allelic loss, and some are involved in homozygous deletions (1, 23, 43). To analyze whether any homozygous deletions or rearrangements of the FEZ1 gene occurred in tumor cells, we studied the FEZ1 gene locus by Southern blotting with the FEZ1 ORF probe in 4 esophageal, 3 prostate, and 11 breast cancer cell lines. The results showed 1 breast cancer cell line with a single rearranged FEZ1 band and loss of the normal allele (Fig. 3D). No homozygous deletions were detected in the other 17 cell lines examined, regardless of the presence or absence of FEZ1 expression. These data suggest that, although LOH in the genomic region around the D8S261 locus, as well as at the FEZ1 gene locus, is a common alteration, homozygous deletions of this gene ORF region are infrequent in tumors. The major mechanism of FEZ1 inactivation may be because of “two-hit” events including allelic loss and point mutations and, possibly, allele loss plus shut-down transcription.
The genomic analysis of region 8p22, a region that is lost preferentially in esophageal squamous cell cancer, as well as in many other tumors, resulted in the identification of FEZ1, a gene encoding a leucine-zipper protein with similarity to the cAMP-responsive Atf-5 DNA-binding protein (44, 45). FEZ1 expression was undetectable in more than 60% of cancers. We detected missense mutations in two primary esophageal cancers and a nonsense mutation in a prostate cancer cell line. These data suggest the FEZ1 tumor suppressor gene candidacy, and that its loss of function plays a role in the development of prostate, breast, and esophagus cancers, and, perhaps, of many other malignancies with alteration at 8p22.
Acknowledgments
We thank Bernadette Mandes and Jean Letofsky for excellent technical assistance. We are grateful to Dr. Kay Huebner for critical reading of the contents of the manuscript. This work was supported in part by U.S. Public Health Service Grants CA39860, CA51083, and CA56336 from the National Cancer Institute. H.I. is an awardee from the Naito Foundation, Japan.
ABBREVIATIONS
- LOH
loss of heterozygosity
- YAC
yeast artificial chromosome
- BAC
bacterial artificial chromosome
- RT
reverse transcription
Footnotes
References
- 1.Cavenee W, Dryja T P, Phillips R A, Benedict W F, Godbout R, Gallie B L, Murphree A L, Strong L C, White R L. Nature (London) 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 2.Lasko D, Cavenee W. Annu Rev Genet. 1991;25:281–314. doi: 10.1146/annurev.ge.25.120191.001433. [DOI] [PubMed] [Google Scholar]
- 3.Weinberg R. Science. 1991;254:1138–1146. doi: 10.1126/science.1659741. [DOI] [PubMed] [Google Scholar]
- 4.Knudson A. Proc Natl Acad Sci USA. 1993;90:10914–10921. doi: 10.1073/pnas.90.23.10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nowell P. Adv Cancer Res. 1993;62:1–17. doi: 10.1016/s0065-230x(08)60313-9. [DOI] [PubMed] [Google Scholar]
- 6.Kagan J, Stein J, Babaian R J, Joe Y-S, Pisters L L, Glassman A B, von Eschenbach A C, Troncoso P. Oncogene. 1995;11:2121–2126. [PubMed] [Google Scholar]
- 7.Macoska J, Trybus T M, Benson P D, Sakr W A, Grignon D J, Wojno K D, Pietruk T, Powell I J. Cancer Res. 1995;55:5390–5395. [PubMed] [Google Scholar]
- 8.Anbazhagan R, Fujii H, Gabrielson E. Am J Pathol. 1998;152:815–819. [PMC free article] [PubMed] [Google Scholar]
- 9.El-Naggar A, Coombes M M, Batsakis J G, Hong W K, Goepfert H, Kagan J. Oncogene. 1998;16:2983–2987. doi: 10.1038/sj.onc.1201808. [DOI] [PubMed] [Google Scholar]
- 10.Sunwoo J, Holt M S, Radford D M, Deeker C, Scholnick S B. Genes Chromosomes Cancer. 1996;16:164–169. doi: 10.1002/(SICI)1098-2264(199607)16:3<164::AID-GCC2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 11.Wu C, Roz L, Sloan P, Read A P, Holland S, Porter S, Scully C, Speight P M, Thakker N. Genes Chromosomes Cancer. 1997;20:347–353. doi: 10.1002/(sici)1098-2264(199712)20:4<347::aid-gcc5>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 12.Wagner U, Bubendorf L, Gasser T C, Moch H, Gorog J P, Richter J, Mihatsch M J, Waldman F M, Sauter G. Am J Pathol. 1997;151:753–759. [PMC free article] [PubMed] [Google Scholar]
- 13.Boige V, Laurent-Puig P, Fouchet P, Flejou J F, Monges G, Bedossa P, Bioulac-Sage P, Capron F, Schmitz A, Olschwang S, Thomas G. Cancer Res. 1997;57:1986–1990. [PubMed] [Google Scholar]
- 14.Takeuchi S, Bartram C R, Wada M, Reiter A, Hatta Y, Seriu T, Lee E, Miller C W, Miyoshi I, Koeffler H P. Cancer Res. 1995;55:5377–5382. [PubMed] [Google Scholar]
- 15.Kerangueven F, Noguchi T, Coulier F, Allione F, Wargniez V, Simony-Lafontaine J, Longy M, Jacquemier J, Sobol H, Eisinger F, Birnbaum D. Cancer Res. 1997;57:5469–5474. [PubMed] [Google Scholar]
- 16.Yaremko M, Recant W M, Westbrook C A. Genes Chromosomes Cancer. 1995;13:186–191. doi: 10.1002/gcc.2870130308. [DOI] [PubMed] [Google Scholar]
- 17.Yaremko M, Kutza C, Lyzak J, Mick R, Recant W M, Westbrook C A. Genes Chromosomes Cancer. 1996;16:189–195. doi: 10.1002/(SICI)1098-2264(199607)16:3<189::AID-GCC6>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 18.Jenkins R, Takahashi S, Delacey K, Bergstralh E, Lieber M. Genes Chromosomes Cancer. 1998;21:131–143. [PubMed] [Google Scholar]
- 19.Gustafson C, Wilson P J, Lukesis R, Baker E, Woollatt E, Annab L, Hawke L, Barret J C, Chenevix-Trench G. Cancer Res. 1996;56:5238–5245. [PubMed] [Google Scholar]
- 20.Ichikawa T, Nihei N, Suzuki H, Oshimura M, Emi M, Nakamura Y, Hayata I, Isaacs J T, Shimazaki J. Cancer Res. 1994;54:2299–2302. [PubMed] [Google Scholar]
- 21.Kuramochi H, Ichikawa T, Nihei N, Kawana Y, Suzuki H, Scalken J A, Takeichi M, Nagafuchi A, Ito H, Shimazaki J. Prostate. 1997;31:14–20. doi: 10.1002/(sici)1097-0045(19970401)31:1<14::aid-pros3>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 22.Nihei H, Ichikawa T, Kawana Y, Kuramochi H, Kugoh H, Oshimura M, Hayata I, Shimazaki J, Ito H. Genes Chromosomes Cancer. 1996;17:260–268. doi: 10.1002/(SICI)1098-2264(199612)17:4<260::AID-GCC8>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 23.Bookstein, R., Bova, G. S., MacGrogan, D., Levy, A. & Isaacs, W. B. (1997) Br. J. Urol.79, Suppl. 1, 28–36. [DOI] [PubMed]
- 24.Bova G, MacGrogan D, Levy A, Pin S S, Bookstein R, Isaacs W B. Genomics. 1996;35:46–54. doi: 10.1006/geno.1996.0321. [DOI] [PubMed] [Google Scholar]
- 25.Cher M, MacGrogan D, Bookstein R, Brown J A, Jenkins R B, Jensen R H. Genes Chromosomes Cancer. 1994;11:153–162. doi: 10.1002/gcc.2870110304. [DOI] [PubMed] [Google Scholar]
- 26.MacGrogan D, Levy A, Bova G S, Isaacs W B, Bookstein R. Genomics. 1996;35:55–65. doi: 10.1006/geno.1996.0322. [DOI] [PubMed] [Google Scholar]
- 27.Fujiwara Y, Ohta H, Kuroki T, Koyama K, Tsuchiya E, Monden M, Nakamura Y. Oncogene. 1995;10:891–895. [PubMed] [Google Scholar]
- 28.Komiya A, Suzuki H, Ueda T, Aida S, Ito N, Shiraishi T, Yatani R, Emi M, Yasuda K, Shimazaki J, Ito H. Jpn J Cancer Res. 1997;88:389–393. doi: 10.1111/j.1349-7006.1997.tb00394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubbard A, Harrison D J, Moyes C, Mylie A H, Cunningham C, Mannion E, Smith C A D. Gut. 1997;41:229–234. doi: 10.1136/gut.41.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matas N, Thygesen P, Stacey M, Risch A, Sim E. Cytogenet Cell Genet. 1997;77:290–295. doi: 10.1159/000134601. [DOI] [PubMed] [Google Scholar]
- 31.Hansson L E, Sparen P, Nyren O. Int J Cancer. 1993;54:402–407. doi: 10.1002/ijc.2910540309. [DOI] [PubMed] [Google Scholar]
- 32.Ozols R. Curr Problems Cancer. 1994;XVIII:191–246. [Google Scholar]
- 33.Ribeiro U, Jr, Posner M C, Safatle-Ribeiro A V, Reynolds J C. Br J Surg. 1996;83:1174–1185. [PubMed] [Google Scholar]
- 34.Nishihira T, Katayama M, Hashimoto Y, Akaishi T. Atlas of Human Tumor Cell Lines. New York: Academic; 1994. pp. 269–285. [Google Scholar]
- 35.Niederacher D, Picard F, van Roeyen C, An H-X, Bender H G, Beckmann M W. Genes Chromosomes Cancer. 1997;18:181–192. doi: 10.1002/(sici)1098-2264(199703)18:3<181::aid-gcc5>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 36.Ausubel F, Brent R, Kingston R E, Moor D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley-Interscience; 1989. pp. 2.5.1–2.6.7. [Google Scholar]
- 37.Bookstein R, Levy A, MacGrogan D, Lewis T B, Weisenbach J, O’Connel P, Leach R J. Genomics. 1994;24:317–323. doi: 10.1006/geno.1994.1622. [DOI] [PubMed] [Google Scholar]
- 38.Akiyama N, Sasaki H, Ishizuka T, Kishi T, Sakamoto H, Onda M, Hirai H, Yazaki Y, Sugimura T, Terada M. Cancer Res. 1997;57:3548–3553. [PubMed] [Google Scholar]
- 39.Elvin P, Butler R, Hedge P J. In: Techniques for the Analysis of Complex Genomes. Anand R, editor. New York: Academic; 1992. pp. 155–171. [Google Scholar]
- 40.Inoue H, Ishii H, Alder H, Snyder E, Druck T, Huebner K, Croce C M. Proc Natl Acad Sci USA. 1997;94:14584–14589. doi: 10.1073/pnas.94.26.14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crick F. Science. 1979;204:264–271. doi: 10.1126/science.373120. [DOI] [PubMed] [Google Scholar]
- 42.Perry, R. (1981) J. Cell. Biol.91, Suppl., 28s–38s. [DOI] [PMC free article] [PubMed]
- 43.Ohta M, Inoue H, Cotticelli M G, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, et al. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 44.Cho-Chung Y, Pepe S, Clair T, Budillon A, Nesterova M. Cri Rev Oncol Hematol. 1995;21:33–61. doi: 10.1016/1040-8428(94)00166-9. [DOI] [PubMed] [Google Scholar]
- 45.Sassone-Corsi P. Annu Rev Cell Dev Biol. 1995;11:355–377. doi: 10.1146/annurev.cb.11.110195.002035. [DOI] [PubMed] [Google Scholar]