Abstract
The function(s) of the genes (PKD1 and PKD2) responsible for the majority of cases of autosomal dominant polycystic kidney disease is unknown. While PKD1 encodes a large integral membrane protein containing several structural motifs found in known proteins involved in cell–cell or cell–matrix interactions, PKD2 has homology to PKD1 and the major subunit of the voltage-activated Ca2+ channels. We now describe sequence homology between PKD2 and various members of the mammalian transient receptor potential channel (TRPC) proteins, thought to be activated by G protein-coupled receptor activation and/or depletion of internal Ca2+ stores. We show that PKD2 can directly associate with TRPC1 but not TRPC3 in transfected cells and in vitro. This association is mediated by two distinct domains in PKD2. One domain involves a minimal region of 73 amino acids in the C-terminal cytoplasmic tail of PKD2 shown previously to constitute an interacting domain with PKD1. However, distinct residues within this region mediate specific interactions with TRPC1 or PKD1. The C-terminal domain is sufficient but not necessary for the PKD2–TRPC1 association. A more N-terminal domain located within transmembrane segments S2 and S5, including a putative pore helical region between S5 and S6, is also responsible for the association. Given the ability of the TRPC to form functional homo- and heteromultimeric complexes, these data provide evidence that PKD2 may be functionally related to TRPC proteins and suggest a possible role of PKD2 in modulating Ca2+ entry in response to G protein-coupled receptor activation and/or store depletion.
Autosomal dominant polycystic kidney disease (ADPKD) is one of the most common genetic diseases, affecting almost one in every thousand Americans (1). Affected individuals can develop kidney and liver cysts, and half of this group manifest end-stage renal failure by age 60. Other affected organs include the pancreas, spleen, and heart, and 2–5% of ADPKD patients also develop cerebral aneurysms (1). By means of positional cloning, two genes, polycystic kidney disease-1 and -2 (PKD1 and PKD2) were identified as candidates for the disease, with mutations in these genes segregating with disease phenotype (2–4).
PKD1 is a large membrane protein with a predicted size of ≈462 kDa. Its extracellular domain contains regions with significant homology to several membrane proteins involved in cell–cell and/or cell–matrix interactions (5, 6). The rest of the molecule is thought to span the membrane 11 times, ending in a relatively short cytoplasmic tail that had been shown to interact with PKD2, the gene product of the second gene found to be responsible for ADPKD (7, 8), and also to bind and activate heterotrimeric Gi/Go proteins in vitro (9).
PKD2 was cloned independently of PKD1 and found to encode an integral membrane protein of 968 amino acids, predicted to span the plasma membrane six times (S1–S6) with its N and C termini placed in the cytoplasm (4). The C-terminal cytoplasmic domain of PKD2 contains a putative EF-hand that partially overlaps with a region that mediates homodimerization, while a more C-terminal region in PKD2 mediates heterodimerization with PKD1 (7, 8). Originally, PKD2 was reported to have homology to a related region in PKD1 and the α1 subunit of the voltage-activated Ca2+ or Na+ channels (4). We now show that it also has considerable homology to the transient receptor potential (TRP) channels (TRPCs). Interestingly, the TRP-homologous region in PKD2 is also homologous to human (4), Caenorhabditis elegans, and Fugu rubripes homologues of PKD1 (5).
Vertebrate TRP genes (TRPC1–7) are mammalian homologues of the Drosophila trp and trp-like (trpl) and are believed to encode putative six membrane-spanning Ca2+ channels activated by depletion of Ca2+ stores (10–13). However, at the present time, it is unclear that all mammalian TRPC proteins form store-operated channels (14). Although there is evidence that TRPC1 (15), TRPC4 (16), and TRPC5 (17) may encode some forms of store-operated channels, functional data pertaining to TRPC6 (18) and Drosophila TRPL (19, 20) do not support this hypothesis. Data in regard to a direct role of TRPC3 in regulating Ca2+ entry in response to store depletion are still evolving (21–23). Such complexity may be explained by the idea that individual TRPC proteins may differentially associate with additional proteins to produce specific functions. This proposal is reasonable because both invertebrate and vertebrate TRP peptides were found to assemble in functional homo- and heteromultimeric complexes (24), raising the possibility that an appropriate combination of TRPCs and/or TRP-related proteins may be required to reveal the true functional properties of these channels.
The fact that PKD2 shared similar topology and had homology with the TRPC proteins over a region that is conserved between PKD2 and PKD1 suggested to us that PKD2 is structurally related to TRPC proteins. We evaluated the hypothesis that PKD2 is functionally related to the TRPCs by testing a possible interaction of PKD2 and the TRPCs. Indeed, PKD2 was found to associate with a ubiquitous member of the mammalian TRPC family, TRPC1, but not with TRPC3. These results point to a specific functional role for PKD2 and also show that individual TRPCs can differentially associate with related proteins. The composition and stoichiometry of these complexes may account for the specific functions of various members of the TRPC family.
MATERIALS AND METHODS
Plasmids and Constructs.
An expression vector containing full-length PKD2 was constructed in pCDNA3 (Invitrogen) by assembling the coding region of PKD2 from clones K1–1 from S. Somlo (Albert Einstein College of Medicine, Bronx, NY) and yj63h09 (Genome Systems). Hemagglutinin epitope-tagged PKD2 (HA-PKD2) was constructed by inserting a synthetic DNA linker encoding the HA epitope (VYPYDVPDYA) in the extracellular region connecting S1–S2 flanked by glycine and serine (GS) residues as follows: G329S330-HA-G341S342C343. Truncation mutants of HA-PKD2 were made by site-directed mutagenesis (25). Full-length calmodulin (CaM) was PCR-amplified from a human brain cDNA library and cloned into pCDNA3. Full-length myc-tagged TRPC1 (M-TRPC1) or myc-tagged TRPC3 (TRPC3-M) tagged at the N or C terminus, respectively, with the myc epitope or the short form of TRPC1 FLAG tagged at its N terminus (F-TRPC1) were obtained from C. Montell (Johns Hopkins Medical School, Baltimore). The α1c subunit of the cardiac voltage-activated Ca2+ channel was obtained from E. Perez-Reyes (Loyola University, Chicago) (26). The myc-tagged form of α1c (α1c-M), was generated by adding the myc epitope at its C terminus by PCR. The Ig.7-PKD2/I679-V968 and sIg.7-PKD2/Q743-E871 constructs were as described (8). To generate fusion constructs with the LexA DNA-binding domain, the C-terminal cytoplasmic region of TRPC1 (D639-S750) or the C terminus of PKD1 (P4124-T4303) was subcloned into the pBHA vector (obtained from M. Sheng, Massachusetts General Hospital, Boston). Progressive N- or C-terminal deletions of the C-terminal cytoplasmic region of PKD2 were subcloned into pADGAL4 (Stratagene) to generate fusions with the transactivation domain of GAL4. Single amino acid substitutions and conversions to stop codons were made by site-directed mutagenesis (25). To generate fusions with glutathione S-transferase (GST), we used a modified version of pGEX-3X (Pharmacia), pGEX-3XL, in which we subcloned cDNA inserts encoding various portions of TRPC1. All constructs that involved PCR were verified by DNA sequencing.
Transfections, Immunoprecipitations, and Immunoblotting.
Human embryonic kidney cells (HEK293T) cells were transfected with 20 μg of total DNA of various combinations of expression plasmids in 10-cm plates by calcium phosphate, and coimmunoprecipitations were done essentially as described (8) with the following modifications. Briefly, cells were lysed in 1 ml/plate of immunoprecipitation buffer (IP-500; 1% Triton X-100/500 mM NaCl/10 mM Tris⋅Cl, pH 7.5/1 mM EDTA/1 mM EGTA/0.2 mM sodium vanadate/0.2 mM phenylmethylsulfonyl fluoride/0.5% Nonidet P-40/10% sucrose containing 1 μg/ml aprotinin, leupeptin, and pepstatin. Lysates were cleared by ultracentrifugation at 100,000 × g for 45 min to pellet partially solubilized microsomes, and 500 μl of cleared lysates were incubated with 10 μg of monoclonal α-HA antibody (mouse IgG2b, Boehringer Mannheim) in a total volume of 1 ml of IP-500 for 2 hr at 4°C. Immunocomplexes were captured with 20 μl of 30% protein A beads for 1 hr at 4°C. Beads were washed four times with IP-500 and immunocomplexes were determined by immunoblotting. When sIg.7 fusions were used, cells were lysed in IP-150 (same as IP-500 except that the final concentration of NaCl was adjusted to 150 mM), and 200 μl of cleared lysates (21,000 × g) was immunoprecipitated directly with 25 μl of 30% protein A beads. Beads were washed three times with IP-150 and immunocomplexes were determined by immunoblotting. Blots were probed with α-HA rabbit polyclonal antibody (Santa Cruz Biotechnology) at 1:400 dilution, α-myc rabbit polyclonal antibody (Santa Cruz Biotechnology) at 1:500 dilution, or α-FLAG rabbit polyclonal antibody (Zymed) at 1:1000 dilution in TTBS (100 mM NaCl/10 mM Tris⋅Cl, pH 7.5/0.2% Tween 20). Bound antibodies were detected by chemiluminescence (Pierce).
Double Immunofluorescence.
HEK293T cells were transfected with 1 μg of HA-PKD2 plus 0.4 μg of F-TRPC1 or M-TRPC1 or 0.1 μg of TRPC3-M by calcium phosphate in 6-well plates. Eight hours after transfection, cells were seeded on coverslips precoated with fibronectin (50 μg/ml), and after 6 hr they were fixed with acetone/methanol. Cells were incubated with a mixture of α-HA (mouse mAb at 10 μg/ml, Boehringer Mannheim) and α-FLAG (rabbit polyclonal antibody at 10 μg/ml, Santa Cruz Biotechnology) antibodies or α-myc (rabbit polyclonal antibody at 10 μg/ml, Santa Cruz Biotechnology) antibody in a total volume of 400 μl of PBS per well for 20 min. After two washes with PBS, cells were incubated with secondary antibodies (rhodamine-conjugated anti-mouse at 1:500 and FITC-conjugated anti-rabbit at 1:200 to detect myc-tagged proteins or 1:300 to detect FLAG-tagged proteins) for 20 min. Cells were washed twice with PBS, and slides were mounted with ProLong Antifade kit (Molecular Probes) and processed for confocal microscopy using a Bio-Rad Confocal Laser Scanning microscope. Superimposed images were obtained by using photoshop (Adobe Systems, Mountain View, CA).
Generation of Fusion Proteins and in Vitro Binding Assays.
Two GST-fusion proteins containing cytoplasmic region D639–S750 or S662–S750 of TRPC1 were purified from BL21 bacterial cells. Specifically, 25-ml portions of bacterial cultures were induced with 200 μM isopropyl β-d-thiogalactoside for 3 hr at 30°C, and recombinant proteins were solubilized in 2 ml of IP-500. Proteins were purified from the soluble fraction with 50 μl of Tris-equilibrated 50% glutathione-Sepharose 4B (Pharmacia) slurry. Beads were washed three times with IP-500 and resuspended in 700 μl of the same buffer. The slurry (10 μl) containing the bound fusion proteins along with 1 or 5 μg of purified GST (Santa Cruz Biotechnology) was separated by SDS/12% polyacrylamide gel. The gel was analyzed by Coomassie brilliant blue to estimate the amount of recombinant GST fusions bound to the beads. It was estimated that ≈10 μg of GST or 4 μg of GST-TRPC1/D639–S750 or GST-TRPC1/S662–S750 was present in 10 μl of each sample. GST (10 μg) or each of the immobilized fusion proteins (4 μg) was incubated with the in vitro transcription–translation reaction mixtures (5 μl; TNT, Promega) containing 35S-PKD2 (residues I679–V968) plus 5 μl of 35S-CaM in 500 μl of IP-500 at 4°C, overnight. Beads were washed five times with IP-500 and bound proteins were eluted in 25 μl of Laemmli buffer (27). The eluted proteins (10 μl) or 1:10 dilution of the TNT reactions (amount of input radioactivity) were separated in an SDS/12% polyacrylamide gel. The gel was dried and exposed to x-ray film for 6 hr.
Yeast Two-Hybrid Assay.
Yeast transformations were performed as described (8). Briefly, 0.2 μg of each plasmid (LexA DNA-binding domain or GAL4 transactivation domain fusion) was used to simultaneously transform the L40 yeast strain that harbors both his and lacZ reporters under the control of LexA binding sites (28). Transformants were plated on media lacking tryptophan and leucine or tryptophan, leucine, and histidine plus 5 mM aminotriazole. Growth on media lacking tryptophan and leucine secured the presence of both plasmids independently of protein–protein interactions and further eliminated the possibility of false negatives. Selection on media lacking tryptophan, leucine, and histidine plus 5 mM aminotriazole was used to detect potential interactions. Cells surviving on these plates were further screened for lacZ+ phenotypes by a filter assay (29). Lifted colonies were scored for lacZ+ phenotypes by detection of blue color in the presence of 5-bromo-4-chloro-3-indolyl β-d-galactoside after incubation at room temperature for 4 hr.
RESULTS
Sequence Similarities Between Members of the PKD2 and TRP Families.
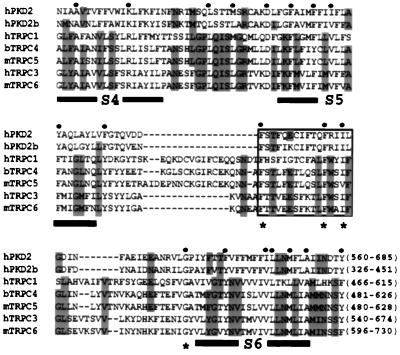
A profile search (prosite, profilescan search) in PKD2 and its homologue PKD2b (as we have named this homologue) or PKD2L (30) revealed the presence of a TRP-like pattern (Fig. 1) and also a channel pore-forming pattern seen in a large number of the pore-forming subunits of Ca2+ and Na+ channels but not K+ channels. The TRP homologous region included the S5 region, the linker between S5 and S6, and the S6 domain and is highly significant in channel function, as it is believed to constitute the ion pore (31, 32). Interestingly, amino acids denoted with an asterisk were found within a putative helical region (see boxed area in Fig. 1) (30) and were conserved between all mammalian TRP proteins (18) and PKD2 or PKD2b. These observations collectively suggest that PKD2 may be structurally related to TRPCs.
Figure 1.
Sequence alignment of the TRP-like regions in human PKD2 (hPKD2, GenBank accession no. U50928), hPKD2b (AF092170), hTRPC1 (Z73903), bovine TRPC4 (bTRPC4, X99792) mouse TRPC5 (mTRPC5, AJ006204), hTRPC3 (Y13758), and mTRPC6 (U49069). The first and last amino acids of each sequence, numbered according to their corresponding cDNAs, are shown in the end of each sequence. Identical or similar substitutions (similar substitutions are grouped as follows: F and Y; I and V; R and K; L and M; or N, D, Q, and E) conserved in five or seven sequences are shown in gray or by a dot on top of the sequences, respectively. Low consensus shown in gray allows the identification of similar residues within the TRP family. Alignment was done according to hierarchical clustering using a blosum62 scoring table (35). Amino acid residues corresponding to putative TM segments (S3–S6) are underlined, and the boxed area shows a putative pore helical region (30). Asterisks denote conserved residues that may participate in the formation of the ion pore.
Specific in Vivo Interaction of PKD2 and TRPC1.
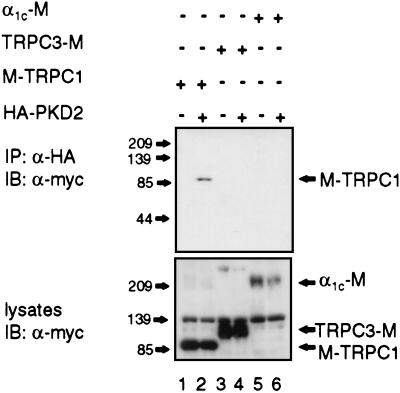
Because TRPC1 and TRPC3 were found to form a heteromultimeric complex in transiently transfected cells (24), we tested whether PKD2 can interact with TRPC1 or TRPC3. We also included the α1c subunit of the cardiac voltage-activated Ca2+ channel (26) because PKD2 was originally reported to have the highest homology to this subfamily of channel proteins (4). Additionally, this particular subunit was found to be expressed in the kidney (33). M-TRPC1 (34), TRPC3-M (24), or α1c-M subunit (26) was expressed in HEK 293T cells and were tested for association with HA-PKD2 by coimmunoprecipitation assays. HA-PKD2 specifically associated with TRPC1 but not with TRPC3 or α1c (Fig. 2A Upper). Similar results were obtained in a converse experiment: HA-PKD2 was coimmunoprecipitated with M-TRPC1 when α-myc was used (data not shown).
Figure 2.
Coimmunoprecipitation of HA-PKD2 and M-TRPC1. HEK293T cells were cotransfected with the indicated combinations of expression plasmids. HA-PKD2 was immunoprecipitated (IP) with α-HA and immunocomplexes were analyzed by immunoblotting (IB) with α-myc (Upper). Expression levels of myc-tagged fusions (M-TRPC1, TRPC3-M, and α1c-M) in the precleared lysates of HEK293T cells before immunoprecipitation are shown (Lower). The positions of molecular mass markers in kDa are shown (Left).
Colocalization of PKD2 and TRPC1 in Live Cells.
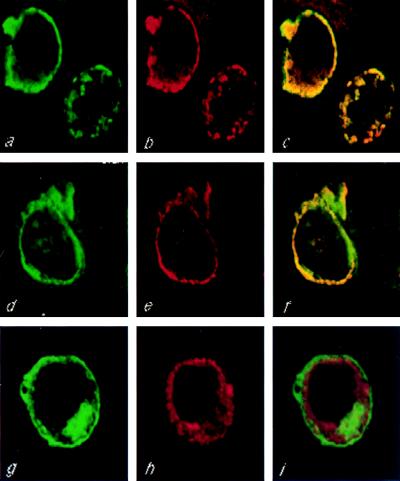
To provide independent evidence that PKD2 and TRPC1 colocalize in live cells, the subcellular distribution of HA-PKD2 and F-TRPC1, M-TRPC1, or TRPC3-M was analyzed by double immunofluorescence in transiently cotransfected cells (Fig. 3). An overlapping subcellular localization was observed between PKD2 and TRPC1 (Fig. 3 c and f) but not TRPC3 (Fig. 3i). These data provide additional evidence that PKD2 specifically interacted with TRPC1 but not TRPC3.
Figure 3.
Colocalization of HA-PKD2 and TRPC1 in live cells. HEK293T cells were cotransfected with HA-PKD2 and F-TRPC1 (a–c), HA-PKD2 and M-TRPC1 (d–f), or HA-PKD2 and TRPC3-M (g–i), and the subcellular distribution of the encoded proteins was determined by double immunofluorescence using confocal microscopy. FLAG- or myc-tagged proteins were stained with a FITC-conjugated secondary antibody, while HA-PKD2 was stained with rhodamine-conjugated secondary antibody. Computerized images of green fluorescein staining corresponding to F-TRPC1, M-TRPC1, or TRPC3-M are shown in a, d, and g, respectively, whereas red rhodamine images corresponding to HA-PKD2 are shown in b, e, and h. Fluorescein and rhodamine merged images corresponding to HA-PKD2 and F-TRPC1, HA-PKD2 and M-TRPC1, or HA-PKD2 and TRPC3-M subcellular distributions are shown in c, f, and i, respectively.
The C-Terminal Cytoplasmic Tail of PKD2 Is Sufficient but Not Necessary to Mediate the Association Between PKD2 and TRPC1.
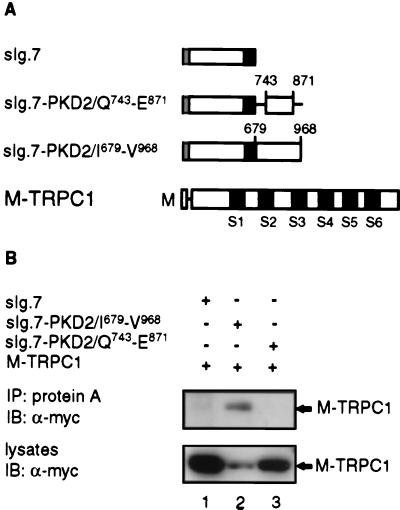
To determine whether the C-terminal cytoplasmic tail of PKD2 was able to interact with TRPC1, we used a heterologous system (8) that would allow us to target the C-terminal tail of PKD2 to the plasma membrane and to test for its association with TRPC1 by coimmunoprecipitation experiments. Portions of the C-terminal tail including amino acid residues I679–V968 or Q743–E871 of PKD2 were fused to an expression cassette bearing the signal sequence of CD5 fused to the CH2–CH3 region of human IgG1 followed by the transmembrane (TM) region of CD7 (Fig. 4A). Both of these constructs, sIg.7-PKD2/I679–V968, which is similar to sIg.7-PKD2 (8), and sIg.7-PKD2/Q743-E871 were shown previously to be efficiently targeted to the membrane (8) and to coimmunoprecipitate with full-length PKD2 (data not shown) because they both retain an active homodimerization domain (8). Fig. 4B shows that sIg.7-PKD2/I679–V968, but not sIg.7-PKD2/Q743–E871, was able to form a complex with M-TRPC1. These data show that the C-terminal region of PKD2 is sufficient to mediate the interaction with TRPC1 and that amino acid residues N terminal to Q743 and/or C terminal to E871 in PKD2 are required for the interaction with TRPC1.
Figure 4.
Coimmunoprecipitation of membrane bound versions of the C-terminal cytoplasmic region of PKD2 and M-TRPC1. (A) Diagrammatic representation of constructs used in (B). The sIg.7 cassette contains the leader sequence of CD5 (░⃞) fused to the CH2-CH3 domain of human IgG1 followed by the TM region of CD7 (■). sIg.7-PKD2/Q743–E871 or sIg.7-PKD2/I679–V968 were made by fusing a subregion (743–871) or the entire C-terminal cytoplasmic tail of PKD2 (679–968) to the C terminus of sIg.7, respectively. Numbering was done according to full-length PKD2 (U50928). Schematic representation of M-TRPC1 including all six TM segments (S1–S6) is also shown. (B) HEK293T cells were cotransfected with sIg.7 (lane 1), sIg.7-PKD2/I679-V968 (lane 2), or sIg.7-PKD2/Q743-E871 (lane 3) and M-TRPC1 (lanes 1–3). sIg.7 fusions were immunoprecipitated (IP) with protein A and immunocomplexes (Upper) or lysates from transiently transfected cells (Lower) were determined by immunoblotting (IB) and probed with α-myc.
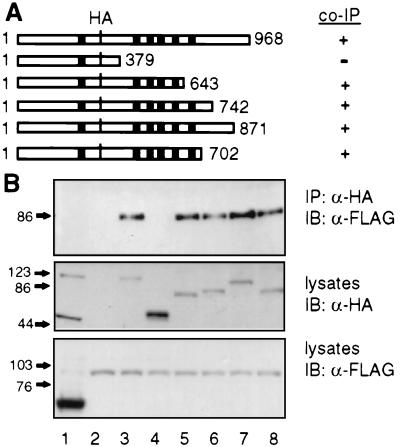
To determine whether the C-terminal tail of PKD2 is also necessary for the interaction with TRPC1, a series of C-terminal truncation mutants of PKD2 were tested for their ability to interact with TRPC1. We found that deletion of the entire C-terminal tail of PKD2, including the S6 (HA-PKDA/1–643) or just the C-terminal cytoplasmic region (HA-PKD2/1–702) did not affect the association with TRPC1, suggesting that an additional domain within the TM domain including S1–S6 was able to mediate the interaction (Fig. 5). Because HA-PKD2/1–379 failed to associate (Fig. 5, lane 4) and HA-PKD2/1–643 was the minimum construct with positive results, we inferred that residues 379–643 constitute an additional interacting domain. Further mutational analysis of this region is needed.
Figure 5.
PKD2-TRPC1 association via a TM region in PKD2. (A) Schematic representation of wild type (1–968; HA-PKD2) or truncation mutants (HA-PKD2/1–379, 1–643, 1–742, 1–871, and 1–702) of HA-PKD2 and summary of results shown in B. Black boxes (■) represent TM segments S1–S6. (B) HEK293T cells were cotransfected with FLAG-tagged bacterial alkaline phosphatase and HA-PKD2 (lane 1), pCDNA3 vector and F-TRPC1 (lane 2), F-TRPC1 and HA-PKD2 (lane 3), HA-PKD2/1–379 (lane 4), HA-PKD2/1–643 (lane 5), HA-PKD2/1–742 (lane 6), HA-PKD2/1–871 (lane 7), or HA-PKD2/1–702 (lane 8). Cells were lysed in IP-500 and HA-tagged proteins were immunoprecipitated (IP) with α-HA. Immunocomplexes were determined by immunoblotting (IB) with α-FLAG (Upper). Expression levels of HA- or FLAG-tagged proteins are shown (Middle or Lower, respectively).
Association of PKD2 and TRPC1 Through Their C-Terminal Tails.
We focused on the C-terminal tail of PKD2 because it was shown previously that it contains an interacting domain with PKD1 (7, 8) and also an EF hand that partially constitutes a homodimerization domain (8). The ability of the C-terminal tails of PKD2 and TRPC1 to interact was determined by two independent assays: GST column binding assays and the yeast two-hybrid assay.
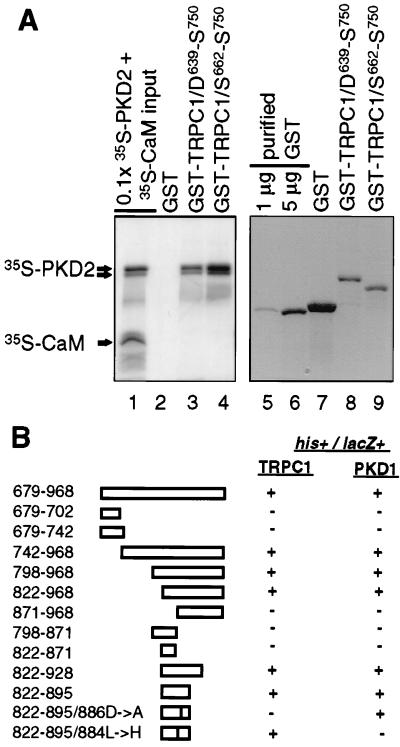
The C-terminal region of TRPC1 (15) (D639–S750) or a subregion containing amino acids S662–S750 was fused to GST and tested for its direct interaction with 35S-labeled probes including the entire cytoplasmic region of PKD2 or full-length human CaM. We chose CaM because, although it has been reported to bind to the C-terminal tails of both Drosophila TRP and TRPL (36, 37), it is not known to interact with mammalian TRPC proteins. Fig. 6A shows that 35S-PKD2 bound to GST-TRPC1/D639–S750 and GST-TRPC1/S662–S750 but not to GST alone. Because CaM did not interact with TRPC1 when added together with 35S-PKD2 (Fig. 6A), it served as an appropriate internal negative control and further supported the specificity of these associations. In addition to these negative controls, the N-terminal region of TRPC1 and the N- or C-terminal tail of TRPC3 were also expressed as GST fusions and tested for direct association with 35S-PKD2. We could not obtain binding between these regions (data not shown). Based on these results, we concluded that region S639–S750 in TRPC1 is sufficient to mediate a specific and direct interaction with PKD2.
Figure 6.
Identification of the C-terminal interacting domain in PKD2. (A) Direct interaction between the C-terminal tail of PKD2 and TRPC1. In vitro column binding of 35S-PKD2 and 35S-CaM to GST-TRPC1/D639–S750 or GST-TRPC1/S662–S750 (lanes 1–4). Lane 1 shows 0.1× the input amount of 35S-PKD2 and 35S-CaM. Bound 35S-PKD2 and 35S-CaM to GST, GST-TRPC1/D639-S750, or GST-TRPC1/S662–S750 is shown in lanes 2–4 and the immobilized amounts of GST, GST-TRPC1/D639–S750, and GST-TRPC1/S662–S750 subjected to in vitro column binding are shown in lanes 7–9. To obtain an estimate of the immobilized amounts of GST, GST-TRPC1/D639–S750, or GST-TRPC1/S662–S750 used in the in vitro column binding assays, 1 or 5 μg of purified GST is shown in lanes 5 and 6. A lower molecular weight band corresponding to a C-terminal proteolytic product of 35S-PKD2 is shown by an arrow below the band corresponding to full-length 35S-PKD2. (B) cDNAs corresponding to the entire C-terminal cytoplasmic region of PKD2, systematic N- or C-terminal deletions of this region (amino acid residues 679–968) or nonconserved substitutions in the region 822–895 were tested for their interaction with the cytoplasmic tail of TRPC1 (D639–S750) or PKD1 (P4124–T4303) by the yeast two-hybrid assay. Positive interactions were scored for both survival in plates lacking histidine (his+) and production of β-galactosidase (lacZ+).
We used the yeast two-hybrid system to provide independent evidence and to define more precisely the domain in PKD2 mediating the interaction between TRPC1 or PKD1. As summarized in Fig. 6B, a minimal region of 73 amino acids (822–895) was sufficient to mediate an interaction with TRPC1 or PKD1. Interestingly, we found that this region (822–895) was consistent with an α-helical conformation according to the Chou–Fasman algorithm (38). To determine the exact amino acid residues in PKD2 responsible for these associations, we introduced several nonconserved substitutions in the C-terminal region of PKD2 from 871 to 895 and evaluated their interaction potential with PKD1 or TRPC1 by using the yeast two-hybrid assay (Fig. 6B). It was found that the L884→H mutation completely inhibited the PKD1–PKD2 interaction, whereas the D886→A mutation inhibited the TRPC1–PKD2 interaction. Thus these data provide additional evidence that PKD2 interacts with TRPC1 or PKD1 and also show that specificity of these interactions is achieved by distinct residues in the C-terminal cytoplasmic region of PKD2.
DISCUSSION
In the present study, we have shown that PKD2 can specifically interact with TRPC1 through two distinct domains. In addition, PKD2 is, to our knowledge, the first non-TRP protein described to specifically interact with TRPC1 but not TRPC3. These data point to a specific role of PKD2 in Ca2+ transport and suggest that specific associations of TRPC proteins with PKD2 and/or PKD2-related molecules may contribute to the functional properties of these channels.
Our initial hypothesis that PKD2 may be interacting with the TRPCs was based on the considerable homology between PKD2 and our newly discovered homologue PKD2b and TRPCs (Fig. 1) and previous studies showing that TRPCs formed functional homo- and heteromultimeric complexes (24). However, PKD2 was originally reported to have the highest homology to the α1 subunit of a large group of voltage-activated Ca2+ and Na+ channels (4). Thus we examined whether PKD2 could associate with the major subunit (α1c) of the cardiac voltage-activated Ca2+ channel (33, 39). We found that PKD2 interacted with TRPC1 but failed to interact not only with α1c but also with an additional member of the TRPC family, TRPC3. These data show that PKD2 specifically interacted with TRPC1 and further eliminated the possibility of a nonspecific interaction with any relatively homologous channel protein. However, the fact that we could not detect an interaction between PKD2 and α1c does not rule out a possible effect of PKD2 on the function of the voltage-activated Ca2+ channels. It is conceivable that PKD2 is interacting with other members of the α1 family or modulates the function of these channels by a mechanism other than physical association.
We have shown that PKD2 interacts with TRPC1 through two distinct domains. Although the C-terminal tails of these peptides engage in a direct physical interaction, the C-terminal tail of PKD2 was not necessary for the interaction with full-length TRPC1 because a more N-terminal region, which was part of the TM domain and included S2–S5 plus part of the linker between S5 and S6, was able to mediate an interaction independently of the C-terminal region. These findings raise two possibilities. First, TRPC1 nonspecifically associates with wild type or truncation mutants despite the stringent immunoprecipitation conditions used or, second, the TM region constitutes a true binding site with functional consequences, particularly because it contains important structural elements involved in channel function. Several lines of evidence support the latter. The TM region of Drosophila TRP coimmunoprecipitated with full-length TRP in transiently transfected HEK293T cells (24). TRPC1 was not able to coimmunoprecipitate with the N-terminal region of PKD2 (Fig. 5, lane 4) or with a subregion of a C-terminal cytoplasmic tail of PKD2 able to mediate homodimerization (Fig. 4, lane 3). Therefore, similarly to Drosophila TRP, PKD2 may associate with TRP peptides through its TM domain to form functional multimeric complexes. Additional regions may further contribute to the overall association.
Because the TM-interacting region lies within the TRP-homologous region in PKD2, which is also conserved in the human, F. rubripes, and C. elegans homologues of PKD1, it is plausible that PKD1 may also associate with TRPC1 and/or TRPC-related proteins through this region. In fact, we have discovered that PKD1 also interacts with multiple members of the TRPC family independently of PKD2, and overexpression of all three proteins does not disturb pairwise interactions (L.T., H. Nomura, M. C. Schneider, G.W., and V.P.S., unpublished work). Thus it is likely that mutations in PKD1 or PKD2 may result in aberrant responsiveness to store depletion that may in turn account for ADPKD. In support of this hypothesis, epithelial cells derived from liver ducts of ADPKD patients showed altered regulation of intracellular Ca2+ in response to the activation of the P2μ purinergic receptor, a G protein-coupled receptor.§
Our results point to a specific function for PKD2 in regulating Ca2+ transport in response to store depletion. This is consistent with the primary structure, subcellular localization of PKD2, and our previous findings that PKD2 interacts with PKD1, a multispanning plasma membrane protein (8). If our hypothesis is proven, the present study will not only provide important insight into the molecular basis of ADPKD but will also help elucidate a poorly characterized pathway mediating a fundamental process in cell biology.
Acknowledgments
We thank Drs. S. Somlo, C. Montell, E. Perez-Reyes, and M. Sheng for plasmids. We also thank Lorenz Sellin for help with confocal microscopy and M. Segal, B. Knebelmann, G. McDonald, W. Schilling, and S. Alper for helpful discussions. This work was supported by Public Health Service Grant DK-09625 (L.T.), MH-01147 (E.K.), a grant from the Polycystic Kidney Research Foundation (G.W.), and seed funds from the Beth Israel Deaconess Medical Center (V.P.S.).
ABBREVIATIONS
- ADPKD
autosomal dominant polycystic kidney disease
- TRP
transient receptor potential
- TRPC
TRP channels
- TM
transmembrane domain (composite region containing all S1–S6)
- HA
hemagglutinin
- HA-PKD2
HA epitope-tagged PKD2
- M-TRPC1
myc-tagged TRPC1
- F-TRPC1
FLAG-tagged TRPC1
- TRPC3-M
myc-tagged TRPC3
- α1c-M
myc-tagged α1c
- CaM
calmodulin
- GST
glutathione S-transferase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF092170).
Jefferson, D. M., Grubman, S. A., Lee, D. W. & Perone, R. D. National Institute of Diabetes and Digestive and Kidney Diseases Workshop on Polycystic Kidney Disease, Sept. 10–11, 1997, Crystal City, VA.
References
- 1.Gabow P A. N Engl J Med. 1993;329:332–342. doi: 10.1056/NEJM199307293290508. [DOI] [PubMed] [Google Scholar]
- 2.The European Polycystic Kidney Disease Consortium. Cell. 1994;77:881–894. doi: 10.1016/0092-8674(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 3.The International Polycystic Kidney Disease Consortium. Cell. 1995;81:289–298. doi: 10.1016/0092-8674(95)90339-9. [DOI] [PubMed] [Google Scholar]
- 4.Mochizuki T, Wu G, Hayashi T, Xenophontos S L, Veldhuisen B, Saris J J, Reynolds D M, Cai Y, Gabow P A, Pierides A, et al. Science. 1996;272:1339–1342. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- 5.Sandford R, Sgotto B, Aparicio S, Brenner S, Vaudin M, Wilson R K, Chissoe S, Pepin K, Bateman A, Chothia C, Hughes J, Harris P. Hum Mol Genet. 1997;6:1483–1489. doi: 10.1093/hmg/6.9.1483. [DOI] [PubMed] [Google Scholar]
- 6.Hughes J, Ward C J, Peral B, Aspinwall R, Clark K, San Millan J L, Gamble V, Harris P C. Nat Genet. 1995;10:151–160. doi: 10.1038/ng0695-151. [DOI] [PubMed] [Google Scholar]
- 7.Qian F, Germino F J, Cai Y, Zhang X, Somlo S, Germino G G. Nat Genet. 1997;16:179–183. doi: 10.1038/ng0697-179. [DOI] [PubMed] [Google Scholar]
- 8.Tsiokas L, Kim E, Arnould T, Sukhatme V P, Walz G. Proc Natl Acad Sci USA. 1997;94:6965–6970. doi: 10.1073/pnas.94.13.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parnell S C, Magenheimer B S, Maser R L, Rankin C A, Smine A, Okamoto T, Calvet J P. Biochem Biophys Res Commun. 1998;251:625–631. doi: 10.1006/bbrc.1998.9514. [DOI] [PubMed] [Google Scholar]
- 10.Minke B, Selinger Z. Mol Neurobiol. 1996;12:163–180. doi: 10.1007/BF02740652. [DOI] [PubMed] [Google Scholar]
- 11.Montell C. Mol Pharmacol. 1997;52:755–763. doi: 10.1124/mol.52.5.755. [DOI] [PubMed] [Google Scholar]
- 12.Birnbaumer L, Zhu X, Jiang M, Boulay G, Peyton M, Vannier B, Brown D, Platano D, Sadeghi H, Stefani E, Birnbaumer M. Proc Natl Acad Sci. USA. 1996;93:15195–15202. doi: 10.1073/pnas.93.26.15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagamine K, Kudoh J, Minoshima S, Kawasaki K, Asakawa S, Ito F, Shimizu N. Genomics. 1998;54:124–131. doi: 10.1006/geno.1998.5551. [DOI] [PubMed] [Google Scholar]
- 14.Clapham D E. Neuron. 1996;16:1069–1072. doi: 10.1016/s0896-6273(00)80132-4. [DOI] [PubMed] [Google Scholar]
- 15.Zitt C, Zobel A, Obukhov A G, Harteneck C, Kalkbrenner F, Luckhoff A, Schultz G. Neuron. 1996;16:1189–1196. doi: 10.1016/s0896-6273(00)80145-2. [DOI] [PubMed] [Google Scholar]
- 16.Philipp S, Cavalie A, Freichel M, Wissenbach U, Zimmer S, Trost C, Marquart A, Murakami M, Flockerzi V. EMBO J. 1996;15:6166–6171. [PMC free article] [PubMed] [Google Scholar]
- 17.Philipp S, Hambrecht J, Braslavski L, Schroth G, Freichel M, Murakami M, Cavalie A, Flockerzi V. EMBO J. 1998;17:4274–4282. doi: 10.1093/emboj/17.15.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boulay G, Zhu X, Peyton M, Jiang M, Hurst R, Stefani E, Birnbaumer L. J Biol Chem. 1997;272:29672–29680. doi: 10.1074/jbc.272.47.29672. [DOI] [PubMed] [Google Scholar]
- 19.Obukhov A G, Harteneck C, Zobel A, Harhammer R, Kalkbrenner F, Leopoldt D, Luckhoff A, Nurnberg B, Schultz G. EMBO J. 1996;15:5833–5838. [PMC free article] [PubMed] [Google Scholar]
- 20.Scott K, Sun Y, Beckingham K, Zuker C S. Cell. 1997;91:375–383. doi: 10.1016/s0092-8674(00)80421-3. [DOI] [PubMed] [Google Scholar]
- 21.Kiselyov K, Xu X, Mozhayeva G, Kuo T, Pessah I, Mignery G, Zhu X, Birnbaumer L, Muallem S. Nature (London) 1998;396:478–482. doi: 10.1038/24890. [DOI] [PubMed] [Google Scholar]
- 22.Zitt C, Obukhov A G, Strubing C, Zobel A, Kalkbrenner F, Luckhoff A, Schultz G. J Cell Biol. 1997;138:1333–1341. doi: 10.1083/jcb.138.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu X, Jiang M, Birnbaumer L. J Biol Chem. 1998;273:133–142. doi: 10.1074/jbc.273.1.133. [DOI] [PubMed] [Google Scholar]
- 24.Xu X Z, Li H S, Guggino W B, Montell C. Cell. 1997;89:1155–1164. doi: 10.1016/s0092-8674(00)80302-5. [DOI] [PubMed] [Google Scholar]
- 25.Higuchi R, Krummel B, Saiki R K. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei X Y, Perez-Reyes E, Lacerda A E, Schuster G, Brown A M, Birnbaumer L. J Biol Chem. 1991;266:21943–21947. [PubMed] [Google Scholar]
- 27.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Vojtek A B, Hollenberg S M, Cooper J A. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 29.Breeden L, Nasmyth K. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 30.Nomura H, Turco A E, Pei Y, Kalaydjieva L, Schiavello T, Weremowicz S, Ji W, Morton C C, Meisler M, Reeders S T, Zhou J. J Biol Chem. 1998;273:25967–25973. doi: 10.1074/jbc.273.40.25967. [DOI] [PubMed] [Google Scholar]
- 31.Catterall W A. Annu Rev Biochem. 1995;64:493–531. doi: 10.1146/annurev.bi.64.070195.002425. [DOI] [PubMed] [Google Scholar]
- 32.Doyle D A, Cabral J M, Pfuetzner R A, Kuo A, Gulbis J M, Cohen S L, Chait B T, MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 33.Yu A S, Hebert S C, Brenner B M, Lytton J. Proc Natl Acad Sci. USA. 1992;89:10494–10498. doi: 10.1073/pnas.89.21.10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wes P D, Chevesich J, Jeromin A, Rosenberg C, Stetten G, Montell C. Proc Natl Acad Sci USA. 1995;92:9652–9656. doi: 10.1073/pnas.92.21.9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chevesich J, Kreuz A J, Montell C. Neuron. 1997;18:95–105. doi: 10.1016/s0896-6273(01)80049-0. [DOI] [PubMed] [Google Scholar]
- 37.Warr C G, Kelly L E. Biochem J. 1996;314:497–503. doi: 10.1042/bj3140497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chou P Y, Fasman G D. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 39.Perez-Reyes E, Schneider T. Kidney Int. 1995;48:1111–1124. doi: 10.1038/ki.1995.395. [DOI] [PubMed] [Google Scholar]