Abstract
Overexpression of the MYC protooncogene has been implicated in the genesis of diverse human tumors. Tumorigenesis induced by MYC has been attributed to sustained effects on proliferation and differentiation. Here we report that MYC may also contribute to tumorigenesis by destabilizing the cellular genome. A transient excess of MYC activity increased tumorigenicity of Rat1A cells by at least 50-fold. The increase persisted for >30 days after the return of MYC activity to normal levels. The brief surfeit of MYC activity was accompanied by evidence of genomic instability, including karyotypic abnormalities, gene amplification, and hypersensitivity to DNA-damaging agents. MYC also induced genomic destabilization in normal human fibroblasts, although these cells did not become tumorigenic. Stimulation of Rat1A cells with MYC accelerated their passage through G1/S. Moreover, MYC could force normal human fibroblasts to transit G1 and S after treatment with N-(phosphonoacetyl)-l-aspartate (PALA) at concentrations that normally lead to arrest in S phase by checkpoint mechanisms. Instead, the cells subsequently appeared to arrest in G2. We suggest that the accelerated passage through G1 was mutagenic but that the effect of MYC permitted a checkpoint response only after G2 had been reached. Thus, MYC may contribute to tumorigenesis through a dominant mutator effect.
Malignant tumors arise from a sequence of events including mutations in protooncogenes and tumor suppressor genes (1). The accretion of these mutations is apparently facilitated by acquired or inherited defects in “guardian” mechanisms that maintain the integrity of the cellular genome (2). There are many examples of these mechanisms. DNA damaged by spontaneous errors or by mutagens is actively repaired (3, 4). Various “checkpoint controls” assure proper progression through the cell cycle, reducing the occurrence of spontaneous DNA damage (5, 6). Apoptotic mechanisms ensure that cells with damaged DNA that cannot be repaired are destroyed (7).
The protooncogene MYC has been implicated in the genesis of diverse human tumors (8). The product of MYC is a transcription factor that can elicit either cellular proliferation or apoptosis, depending on physiological conditions (7, 8). The tumorigenic effects of MYC have been generally attributed to sustained effects on cellular proliferation and differentiation (8). MYC may also contribute to tumorigenesis by inducing genomic destabilization (9–12). We report that transient excess of MYC activity can promote tumorigenesis in an immortal rodent cell line and elicit genomic destabilization in both immortal rodent cell lines and in normal human fibroblasts. Our results suggest that MYC may contribute to tumorigenesis by affecting the G1/S checkpoints for control of DNA damage.
METHODS
Cell Lines.
Rat1A is an immortal and pseudodiploid cell line of rat fibroblasts that was recloned before the present work. Normal human fibroblasts (NHF) were obtained from newborn foreskin (kindly provided by Thea Tlsty, University of California, San Francisco). Cells were infected with the pBABE puromycin retrovirus generating Rat1A(BABE) and NHF(BABE) or were infected with pBABE puromycin retrovirus containing MYCER generating Rat1A(MYCER) and NHF(MYCER) (13).
Tumorigenicity Assays.
Rat1A or Rat1A(MYCER) cells were either untreated or exposed to E2 or 4-hydroxytamoxifen (4-OHT) (10 μM) as indicated. The hormone was then withdrawn, and the cells were maintained in culture for the indicated number of days before testing for tumorigenicity. Assays for tumorigenicity were performed by injecting 107 cells subcutaneously into the flanks of BALB/c nude mice (The Jackson Laboratory). Tumors typically appeared after 4 weeks. Monitoring was discontinued after 10 weeks, at which time mice were sacrificed. The frequency of neoplastic cells was estimated by limiting dilution by using the formula: ln(Tf) = −nf, where Tf is the proportion of tumor free mice, n is the number of cells injected into the mice, and f is the frequency of neoplastic cells (14).
Karyotypic Analysis.
To determine chromosome number, metaphase spreads were prepared and evaluated as described (15). To determine mitotic index, metaphase spreads were prepared from cells without prior colchicine treatment. The number of mitotic nuclei were counted.
N-(phosphonoacetyl)-l-aspartate (PALA) Resistance.
PALA resistance was determined at 9-fold LD50 because this dosage routinely selects for genomic amplification of the multifunctional enzyme that contains carbamyl phosphate synthase, aspartate transcarbamylase, and dihydroorotase (CAD) (15). Southern analysis was performed by using a probe for hamster CAD cDNA (15).
Etoposide and UV Sensitivity.
Rat1A(MYCER) or NHF(MYCER) were irradiated with UV (1–20 J/m2) or exposed to etoposide (0.01–10 μg/ml) and then treated with 0.1 μM estrogen (E2). After 5 days, trypan blue-excluding adherent and nonadherent cells were counted.
BrdUrd Incorporation.
Staining with anti-BrdUrd-FITC and propidium iodide were performed as described by the manufacturer (Becton Dickinson).
RESULTS
Transient Excess of MYC Activity Augments Tumorigenicity.
We produced increases in MYC activity by using a chimeric gene (MYCER) in which MYC is fused to the hormone-binding domain of the human estrogen receptor (13). The MYCER protein is active only in the presence of E2 or 4-OHT. If overexpressed at a sufficient level, the activated MYCER protein transforms cells in vitro (13). By withdrawing E2, transformation is reversed.
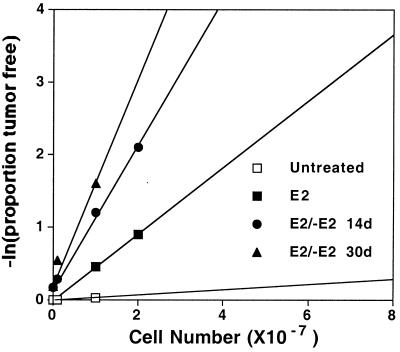
The Rat1A cell line of embryonic fibroblasts is pseudodiploid and morphologically normal, but is readily transformed by MYC in vitro (16). These cells displayed little or no tumorigenicity after cultivation in either the absence or presence of E2 (Table 1). In contrast, Rat1A cells expressing MYCER [Rat1A(MYCER)] became notably tumorigenic in mice after in vitro exposure to E2 or 4-OHT (Table 1, Fig. 1, and data not shown). As expected, the same treatment also elicited full morphological transformation (data not shown, see also ref. 13). We were surprised to find, however, that withdrawal of hormone for periods as long as 30 days did not reverse tumorigenicity (Table 1), even though the cells resumed their normal appearance in vitro and had undergone a minimum of 15 population doublings. Indeed, the fraction of tumorigenic cells appeared to increase up to 50-fold over time subsequent to withdrawal of E2 (Table 1 and Fig. 1). Similar results were obtained with several independent clones of Rat1A(MYCER) cells.
Table 1.
Transient excess of MYC activity elicits tumorigenicity
| Cell line | Treatment, days
|
Tumors*, % | Tumors/Sites† | n | Frequency‡ | |
|---|---|---|---|---|---|---|
| +E2 | −E2 | |||||
| Rat1A | 0 | 0 | 0 | 0/10 | 2 | <1.0 × 10−8 |
| 10 | 0 | 0 | 0/10 | 2 | <1.0 × 10−8 | |
| Rat1A(MYCER) | 0 | 0 | 3 ± 5 | 2/56 | 8 | 3.0 × 10−9 |
| 2 | 0 | 36 ± 9 | 13/34 | 4 | 4.5 × 10−8 | |
| 10 | 0 | 34 ± 10 | 28/72 | 8 | 4.2 × 10−8 | |
| 2 | 2 | 42 | 5/12 | 1 | 6.9 × 10−8 | |
| 10 | 2 | 30 | 3/10 | 1 | 3.6 × 10−8 | |
| 2 | 14 | 64 ± 6 | 14/22 | 2 | 1.0 × 10−7 | |
| 10 | 14 | 60 | 6/10 | 1 | 9.2 × 10−8 | |
| 2 | 21 | 58 ± 16 | 14/24 | 2 | 8.7 × 10−8 | |
| 2 | 30 | 75 | 6/8 | 1 | 1.4 × 10−7 | |
Cells were exposed in vitro to E2 (10 μM) as indicated.
Percent of sites with tumors ±SD.
Data aggregated from individual experiments.
Estimated frequency of neoplastic cells (14).
Figure 1.
Brief excess of MYC activity increases the frequency of tumorigenic cells. Rat1A(MYCER) untreated (□), treated with E2 (10 μM) for 2 days (■), treated with E2 for 2 days followed by 14 days (●) or 30 days (▴) in the absence of hormone treatment. Tumorigenicity assays were performed as described in Table 1.
We conclude that brief exposure to high levels of MYC activity is sufficient to enhance tumorigenicity of Rat1A cells that is stable over many days and many cell divisions. We do not believe that these results can be explained by residual function of MYCER in the absence of E2. First, Rat1A(MYCER) cells that had never been exposed to E2 were only feebly tumorigenic (Table 1). Second, the serum concentration of E2 in mice was measured and found to be 10–20 pm, 1,000th of that required to activate MYCER. Third, the protein expression of MYCER and endogenous MYC were equivalent in the cells before and after induction of tumorigenicity (data not shown). Thus, tumorigenicity cannot be ascribed to occasional clones of cells that produce exceptional amounts of MYC or MYCER. Fourth, cell lines established from the Rat1A(MYCER) tumors were morphologically normal. E2 treatment of these cells elicited morphological transformation in the presence of normal levels of serum and apoptosis in reduced serum (0.5%, data not shown). Fifth, we examined expression of the ornithine decarboxylase gene (ODC), whose transcription is induced by MYC (17). Expression of ODC was undetectable in cells from a tumor generated by 48-hour exposure of Rat1A(MYCER) cells to E2 but subsequently cultivated in reduced serum (0.5%) in the absence of E2 (data not shown). In contrast, ODC protein was readily detectable after treatment of either the tumor cells or of the original Rat1A(MYCER) cells with E2. These diverse observations demonstrate that the activity of MYCER remained conditional and was negligible in the absence of E2.
Transient Excess of MYC Activity Destabilizes the Genome.
The durable tumorigenic phenotype conferred by brief excess of MYC activity prompted us to investigate whether MYC is capable of inducing genomic changes in Rat1A cells. We found that treatment of Rat1A(MYCER) cells with E2 for 2 days produced multiple karyotypic abnormalities, including marked aneuploidy, polycentric chromosomes, double minute chromosomes, and chromosome breaks (Table 2 and Fig. 2). The frequency of chromosomal abnormalities was not increased by prolonging the exposure to E2 (Table 2). A mutation that removed the transcriptional-activation domain of MYC from MYCER (13) eliminated the chromosomal response to E2 by Rat1A cells (data not shown). In addition, E2 had no effect on the karyotype of normal Rat1A cells that contained only the vector used to express MYCER (Table 2 and data not shown). After withdrawal of E2, the karyotype of Rat1A(MYCER) returned virtually to normal: the frequency of aneuploid cells diminished to background levels (Table 2), and neither polycentric nor double minute chromosomes were detected (Table 2). A transient excess of MYC activity also induced genomic destabilization in NIH 3T3 cells (data not shown).
Table 2.
Transient excess of MYC activity induces karyotypic abnormalities
| Cell line | Treatment, days
|
Karyotypic abnormality, %*
|
||||
|---|---|---|---|---|---|---|
| +E2 | −E2 | Aneuploidy† | Polycentric | Double minute | Chromosome break | |
| Rat1A | 0 | 0 | 3 | <1 | <1 | <1 |
| 14 | 0 | 6 | <1 | <1 | <1 | |
| Rat1A(MYCER) | 0 | 0 | 2 | <1 | <1 | <1 |
| 2 | 0 | 22 | 21 | 7 | 2 | |
| 14 | 0 | 20 | 14 | 5 | <1 | |
| 14 | 2 | 19 | 7 | 4 | <1 | |
| 14 | 14 | 5 | <1 | <1 | <1 | |
| Tumor 1 | 0 | 0 | 28 | 13 | <1 | <1 |
| 2 | 0 | 62 | 44 | 5 | 4 | |
| Tumor 2 | 0 | 0 | 8 | 11 | <1 | <1 |
| 2 | 0 | 54 | 22 | <1 | <1 | |
| NHF | 0 | 0 | <1 | <1 | <1 | <1 |
| 2 | 0 | <1 | <1 | <1 | <1 | |
| NHF(MYCER) | 0 | 0 | <1 | <1 | <1 | <1 |
| 2 | 0 | 21 | <1 | <1 | <1 | |
Cells were treated with E2 as described for Table 1. Tumors 1 and 2 were explanted Rat1A(MYCER) tumors propagated in vitro. At least 50 metaphases were analyzed per group.
Percent metaphases with indicated karyotypic abnormality.
Percent metaphases with 50 or more chromosomes.
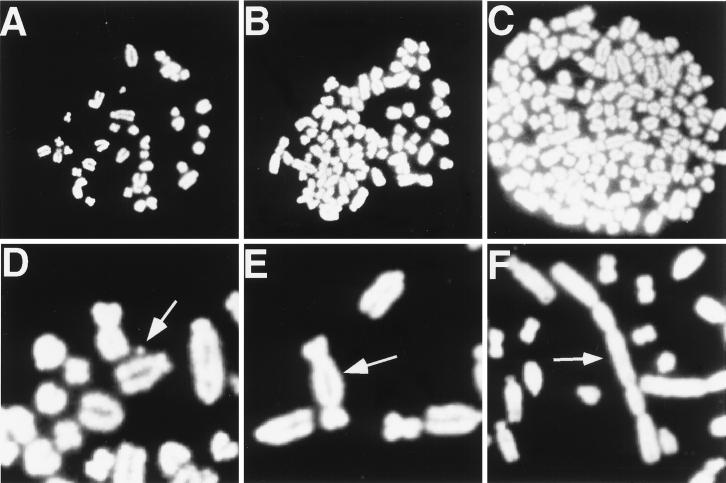
Figure 2.
Excess of MYC activity is associated with karyotypic abnormalities. Cells were treated with E2 (10 μM) for two days, followed by analysis of metaphase spreads (15). Untreated Rat1A(MYCER) cells displayed a normal karyotype (A). Rat1A(MYCER) cells treated with E2 displayed aneuploidy (B), double minute chromosomes (D), and dicentric chromosomes (E). In a separate experiment, Rat1A(MYCER) cells were treated with E2 for 48 hours and then directly injected into nude mice to induce tumors. Explanted tumors were propagated in vitro and found to be euploid, but when treated with E2 exhibited marked aneuploidy (C) and multicentric chromosomes (F).
Cell lines established from explanted Rat1A(MYCER) tumors were euploid (Table 2) but exhibited an exceptional frequency of other karyotypic changes, particularly when exposed to E2 (Table 2, Fig. 2). Presumably, tumorigenic clones have undergone additional genetic events that make them more susceptible to MYC-induced genomic destabilization.
Polycentric chromosomes are thought to be precursors of genomic amplification, and double minute chromosomes are associated with genomic amplification (18). We found that exposure of Rat1A(MYCER) cells to E2 for 2 days increased by 100-fold the frequency of cells resistant to killing by PALA (Table 3). PALA resistance in this assay arose from amplification of the CAD gene, which we documented by using Southern blotting. We found that PALA-resistant cells had 2–10 additional copies of the CAD gene (data not shown). Resistance to PALA was still apparent when the assay was initiated 14 days after withdrawal of E2 (Table 3). Indeed, the fraction of PALA-resistant cells appeared to increase over time up to 200-fold subsequent to withdrawal of E2 (Table 3).
Table 3.
Frequency of PALA resistance is increased by transient excess of MYC activity
| Cell line | Treatment | LD50, μM | Frequency of PALA-resistant clones |
|---|---|---|---|
| Rat1A | None | 30 | <1 × 10−7 |
| E2* | 30 | <1 × 10−7 | |
| Rat1A(MYCER) | None | 45 | <1 × 10−7 |
| E2* | 45 | 9.5 × 10−6 ± 2.4 × 10−6 | |
| E2/None† | ND | 2.1 × 10−5 ± 0.1 × 10−5 |
ND, not determined
E2 (10 μM) for 2 days, then selection in PALA.
E2 (10 μM) for 2 days, then no treatment for 14 days, then selection in PALA.
As a final parameter of MYC’s influence on genomic stability, we analyzed the effect of excess MYC activity on the sensitivity of Rat1A cells to DNA-damaging agents. We found that Rat1A(MYCER) cells were 2-fold more sensitive to UV radiation and 25-fold more sensitive to etoposide in the presence of E2 (Table 4).
Table 4.
Excess of MYC activity increases sensitivity to DNA-damaging agents
| Cell line | Relative sensitivity*
|
|
|---|---|---|
| UV | Etoposide | |
| Rat1A(MYCER) | 1.9 ± 0.1 | 25 ± 7.3 |
| NHF(MYCER) | 4.6 ± 0.4 | not sensitive† |
Results are from three independent experiments, expressed as means ±SD.
LD50 in the absence of E2 divided by the LD50 in the presence of E2.
NHF did not exhibit sensitivity to etoposide at 10 μg/ml.
MYC Induces Genomic Destabilization of NHF.
Because Rat1A cells do not exhibit cellular senescence, they may have genetic alterations that predispose them to the effects of MYC reported here. We therefore turned our attention to NHF. Treatment of NHF(BABE) with E2 produced none of the abnormalities associated with genomic destabilization. In contrast, exposure of NHF that were expressing NHF(MYCER) to E2 produced abundant aneuploidy (Table 2) and a 5-fold-increased sensitivity to UV radiation (Table 4). NHF were insensitive to etoposide even in the presence of excess MYC activity.
E2 treatment of NHF(MYCER) failed to elicit polycentric chromosomes, double minute chromosomes, chromosome breaks or an increase in PALA resistance. These results suggest that an excess of MYC activity may not produce chromosome damage in NHF. We note, however, that normal cells can cease to proliferate in the presence of even a single double-strand DNA break (19), which would make it difficult to detect chromosomal damage with the analyses employed here.
Excess MYC Activity Prevents Cell Cycle Arrest in Response to DNA Damage.
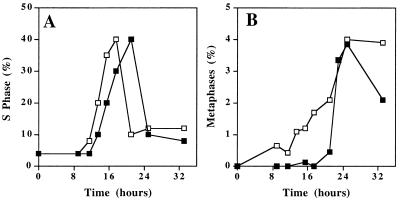
How does MYC induce genomic destabilization? MYC is known to accelerate passage through G1/S in immortal rodent cells (20). Acceleration through the cell cycle in itself may be mutagenic for several reasons (5). We confirmed that excess MYC activity in Rat1A cells accelerated passage through G1/S by 4 hours, as previously documented (Fig. 3A). However, when we analyzed individual cells for entry into metaphase, we found that MYC accelerated the cell cycle by as much as 16 hours (Fig. 3B). Similar results were seen for NHF (data not shown).
Figure 3.
Excess MYC activity accelerates transit through the cell cycle. Serum-starved Rat1A(MYCER) cells were treated with serum (10%, ■) or serum and E2 (1 μM, □). (A) S Phase was determined by measuring the frequency of cells that incorporated BrdUrd. (B) Mitotic index was measured by examining the frequency of mitotic nuclei in metaphase spreads.
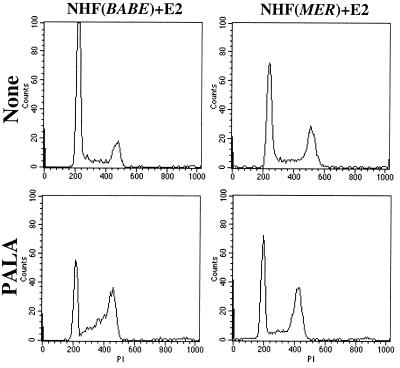
Although acceleration of passage through the cell cycle could itself be mutagenic, we wondered whether MYC might also perturb the ability of cells to adequately respond to conditions that promoted DNA damage. To address this issue, we returned to the examination of NHF, which exhibit cell cycle arrest in response to DNA damage. We exposed NHF to PALA in the presence or absence of excess MYC activity. PALA inhibits CAD, thereby restricting nucleotide pools leading to conditions that promote DNA damage in normal rodent and human cells (15). We exposed NHF to PALA at a concentration that we determined to be 9-fold LD50. As expected, when we treated asynchronously growing populations of NHF, they had difficulty completing transit through the cell cycle, accumulating in S phase (Fig. 4). However, the E2 treatment of NHF(MYCER) suppressed this accumulation in S phase (Fig. 4). Instead, E2-treated NHF(MYCER) accumulated in G2 (Fig. 4). In contrast, E2 treatment of NHF(BABE) cells had no effect. Thus, excess MYC activity may abrogate checkpoints for DNA damage that function during G1/S. As a result, DNA damage accumulates, and this in turn leads to the arrest of cell cycle in G2.
Figure 4.
Excess MYC activity causes inappropriate cell cycle entry. NHF(BABE) or NHF(MYCER) cells were treated with PALA (50 μM) for two days and then analyzed for DNA content by fluorescence-ativated cell sorter (FACS) analysis of propidium iodide stained cells.
DISCUSSION
We conclude that a transient excess of MYC activity can predispose cells to tumorigenesis. Most likely this occurs because the cellular genome has been destabilized, creating a mutator phenotype that increases the frequency of tumors produced by an established line of rodent cells. Tumorigenic cells remained rare (10−7 to 10−8) after destabilization of the genome, perhaps because multiple mutations are required for tumorigenicity even in Rat1A cells. The tumorigenicity of Rat1A(MYCER) cells in which the genome has been destabilized persists for at least 30 days after removal of E2 and the frequency of tumorigenic cells may even increase over that time (Fig. 1). In contrast, cells with karyotypic abnormalities do not persist in the population after E2 removal, perhaps because such cells are killed by apoptosis. Gene amplification persists and may also increase during the two weeks after withdrawal of E2, indicating that genetic alterations do persist after transient surplus of MYC activity (Table 3). Our observations are consistent with several reports that indicate MYC and other oncogenes induce genomic destabilization (9–12, 21–24).
How might a transient surfeit of MYC activity destabilize the genome? First, abbreviation of G1 may be mutagenic (5). Indeed, G1 is curtailed in Rat1A(MYCER) and NHF(MYCER) cells treated with E2 (20). In fact, our data suggest that MYC may accelerate passage through G1 by a greater degree than previously described (Fig. 3). Second, excess activity of MYC forces cells into the cell cycle under deleterious conditions such as those imposed by treating cells with high concentrations of PALA (Fig. 4). This too may destabilize the genome by increasing DNA damage (25). Similar results have been described for MYC’s ability to abrogate the cell cycle arrest of REF52 cells that have been treated with PALA (12). Third, MYC may reduce transcription or inhibit the function of gene products responsible for regulating cell cycle transit (12, 26–28).
Our data suggest that MYC prevents a checkpoint response during G1/S. Instead, a checkpoint response appears only after G2 had been reached (Fig. 4). The strict imposition of G2 arrest in NHF in response to MYC suggests that genomic damage has indeed occurred. Thus, MYC may contribute to tumorigenesis through a dominant mutator effect by abrogating checkpoints during G1/S. As such, MYC exemplifies a postulated category of a dominant-acting oncogene that generates aneuploidy (29).
Lymphomas in which MYC is overexpressed were formerly thought to be euploid. At first glance, this appears to be paradoxical. However, recent analyses with techniques such as comparative genomic hybridization and spectral karyotyping (which possess enhanced resolution over conventional karyotypic analysis) reveal that chromosomal abnormalities are actually common in these tumors (30).
The results presented here provide the first indication that MYC may in some circumstances initiate tumorigenesis by a “hit-and-run” mechanism. In this scenario, there may be no requirement for sustained MYC overexpression once tumor progression has been launched by destabilization of the genome. Thus, there may be tumors in which therapeutic strategies that target the inactivation of MYC will not be effective.
Acknowledgments
This work is dedicated to the memory of Donby Lee. We thank Thea Tlsty for providing NHF and advice on performing PALA assays, Phillip Coffino for providing a rabbit polyconal antibody against ODC protein, Paul Dazin for performing FACS analysis, the members of the Bishop laboratory for generously providing reagents and their many helpful suggestions, and Geoff Wahl, Ira Herskowitz, Thea Tlsty, and Jiyue Zhu for critical review of the manuscript. D.W.F. is a fellow of the Howard Hughes Medical Institute and a recipient of the Lymphoma Research Foundation Fellowship and the Pfizer Postdoctoral Fellowship. Work was supported by funds from the National Institutes of Health (Grant CA 44338) and the G. W. Hooper Research Foundation.
ABBREVIATIONS
- NHF
normal human fibroblasts
- PALA
N-(phosphonoacetyl)-l-aspartate
- CAD
multifunctional enzyme that contains carbamyl phosphate synthase, aspartate transcarbamylase, and dihydroorotase
- E2
estrogen
- 4-OHT
4-hydroxytamoxifen
References
- 1.Bishop J M. Cell. 1991;64:235–248. doi: 10.1016/0092-8674(91)90636-d. [DOI] [PubMed] [Google Scholar]
- 2.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 3.Modrich P. Science. 1994;266:1959–1960. doi: 10.1126/science.7801122. [DOI] [PubMed] [Google Scholar]
- 4.Elledge S J. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 5.Paulovich A G, Toczyski D P, Hartwell L H. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 6.Hartwell L H, Kastan M B. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 7.Evan G I, Brown L, Whyte M, Harrington E. Curr Opin Cell Biol. 1995;7:825–834. doi: 10.1016/0955-0674(95)80066-2. [DOI] [PubMed] [Google Scholar]
- 8.Marcu K B, Bossone S A, Patel A J. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 9.Mai S, Hanley-Hyde J, Fluri M. Oncogene. 1996;12:277–288. [PubMed] [Google Scholar]
- 10.Mai S, Fluri M, Siwarski D, Huppi K. Chromosome Res. 1996;4:365–371. doi: 10.1007/BF02257272. [DOI] [PubMed] [Google Scholar]
- 11.Cerni C, Mougneau E, Zerlin M, Julius M, Marcu K B, Cuzin F. Curr Top Microbiol Immunol. 1986;132:193–201. doi: 10.1007/978-3-642-71562-4_28. [DOI] [PubMed] [Google Scholar]
- 12.Chernova O B, Chernov M V, Ishizaka Y, Agarwal M L, Stark G R. Mol Cell Biol. 1998;18:536–545. doi: 10.1128/mcb.18.1.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eilers M, Picard D, Yamamoto K R, Bishop J M. Nature (London) 1989;340:66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- 14.Fazekas de St G. J Immunol Methods. 1982;49:R11–R23. doi: 10.1016/0022-1759(82)90269-1. [DOI] [PubMed] [Google Scholar]
- 15.Tlsty T D. Methods Mol Genet. 1996;8:388–401. [Google Scholar]
- 16.Small M B, Hay N, Schwab M, Bishop J M. Mol Cell Biol. 1987;7:1638–1645. doi: 10.1128/mcb.7.5.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bello-Fernandez C, Packham G, Cleveland J L. Proc Natl Acad SciUSA. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Windle B, Draper B W, Yin Y X, O’Gorman S, Wahl G M. Genes Dev. 1991;5:160–174. doi: 10.1101/gad.5.2.160. [DOI] [PubMed] [Google Scholar]
- 19.Huang L C, Clarkin K C, Wahl G M. Proc Natl Acad SciUSA. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karn J, Watson J V, Lowe A D, Green S M, Vedeckis W. Oncogene. 1989;4:773–787. [PubMed] [Google Scholar]
- 21.Zhou P, Jiang W, Weghorst C M, Weinstein I B. Cancer Res. 1996;56:36–39. [PubMed] [Google Scholar]
- 22.Denko N C, Giaccia A J, Stringer J R, Stambrook P J. Proc Natl Acad SciUSA. 1994;91:5124–5128. doi: 10.1073/pnas.91.11.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukasawa K, Vande Woude G F. Mol Cell Biol. 1997;17:506–518. doi: 10.1128/mcb.17.1.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mumberg D, Haas K, Möröy T, Niedenthal R, Hegemann J H, Funk M, Müller R. Oncogene. 1996;13:2493–2497. [PubMed] [Google Scholar]
- 25.Paulson T G, Almasan A, Brody L L, Wahl G M. Mol Cell Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermeking H, Funk J O, Reichert M, Ellwart J W, Eick D. Oncogene. 1995;11:1409–1415. [PubMed] [Google Scholar]
- 27.Alevizopoulos K, Vlach J, Hennecke S, Amati B. EMBO J. 1997;16:5322–5333. doi: 10.1093/emboj/16.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marhin W W, Chen S, Facchini L M, Fornace A J, Jr, Penn L Z. Oncogene. 1997;14:2825–2834. doi: 10.1038/sj.onc.1201138. [DOI] [PubMed] [Google Scholar]
- 29.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 30.Coleman A E, Schröck E, Weaver Z, du Manoir S, Yang F, Ferguson-Smith M A, Ried T, Janz S. Cancer Res. 1997;57:4585–4592. [PubMed] [Google Scholar]