Abstract
The DNA mismatch repair (MMR) is a specialized system, highly conserved throughout evolution, involved in the maintenance of genomic integrity. To identify novel human genes that may function in MMR, we employed the yeast interaction trap. Using the MMR protein MLH1 as bait, we cloned MED1. The MED1 protein forms a complex with MLH1, binds to methyl-CpG-containing DNA, has homology to bacterial DNA repair glycosylases/lyases, and displays endonuclease activity. Transfection of a MED1 mutant lacking the methyl-CpG-binding domain (MBD) is associated with microsatellite instability (MSI). These findings suggest that MED1 is a novel human DNA repair protein that may be involved in MMR and, as such, may be a candidate eukaryotic homologue of the bacterial MMR endonuclease, MutH. In addition, these results suggest that cytosine methylation may play a role in human DNA repair.
The DNA mismatch repair (MMR) system plays an important role in reducing mutations and maintaining genomic stability (1). MMR corrects base–base mismatches and short insertion/deletion mispairs, generated as a consequence of DNA replication errors and homologous recombination (2–4). The major MMR pathway in Escherichia coli is the methyl-directed MutHLS system. In this system, the mismatch is detected by MutS; then, after interaction with MutL, the single-strand endonuclease MutH is activated and incises the newly synthesized DNA strand that contains the mutation. After excision of a tract of about 1–2 kb containing the misincorporated base, resynthesis occurs (2–4). MMR components are highly conserved in evolution, and germ-line defects in human homologues of mutS (MSH2 and MSH6) and mutL (MLH1, PMS2, and PMS1) are associated with hereditary nonpolyposis colorectal cancer or Lynch syndrome (5–7). Mutations of MMR genes also are detected in a subset of sporadic cancers with microsatellite instability (MSI) (8–9).
In bacterial MMR, MutH identifies and cleaves the new strand by virtue of the latter’s transient lack of adenine methylation at palindromic d(GATC) sites (2–4). Despite its critical function, MutH homologues have not been identified outside of E. coli and the closely related bacterium Salmonella typhimurium (4). Furthermore, the molecular determinants of strand discrimination in eukaryotic cells, which lack d(GATC) methylation, have remained elusive (2–4). In cell-free reconstituted eukaryotic MMR systems, repair of a mismatched plasmid substrate is directed to the appropriate strand by a single-strand nick located either 5′ or 3′ to the mispair (10, 11). Indeed, it has been proposed that DNA termini generated at the replication fork may provide the strand-targeting signal in vivo (2–4) and that proliferating-cell nuclear antigen (PCNA) might aid in this process by physically linking DNA polymerases and MMR proteins (12). An alternative possibility is that eukaryotic endonucleases may exist that, like MutH, are involved in MMR.
To identify novel human genes that may function in MMR, we conducted a yeast interaction trap screening. This screening led to the identification of MED1, an endonuclease that interacts with MLH1 and appears to have features of a candidate homologue of MutH.
MATERIALS AND METHODS
Yeast Interaction Trap Screening, cDNA Isolation, and Sequence Analysis.
For yeast interaction trap screening (13), the entire MLH1 ORF (codons 1–756) was inserted in pEG202 (13), generating the bait construct pEG202-t-MLH1 as a carboxyl-terminal fusion to LexA. Saccharomyces cerevisiae strain EGY191 was transformed with the bait construct and the lacZ reporter pSH18–34 (13). Yeast cells then were supertransformed with a human fetal brain cDNA library in the vector pJG4–5. This vector directs the synthesis of proteins fused to the B42 transactivator domain under the control of the galactose-inducible GAL1 promoter (13). Approximately 4 × 105 independent transformants were obtained and screened. For selection of positive interactors, supertransformed cells were cultured on Leu (−)/galactose plates. Galactose-specific, Leu (+), lacZ-expressing colonies were isolated. Plasmid DNA was rescued from these colonies and sequenced. These and subsequent sequencing reactions were performed on double-strand DNA with ABI sequencer 377 (Perkin–Elmer). Three additional cDNA lambda libraries from human fetal brain (Stratagene and CLONTECH) and one from the ovarian cancer cell line C200 were screened as described previously (14) by using the entire insert of clone f5 as probe. Sequence assembling and analysis were performed with the gcg software package (Wisconsin Package Version 9.1, GCG). Sequence alignments were displayed with boxshade (http://ulrec3.unil.ch/software/BOX_form.html).
Cell Culture, Expression Constructs, and Transfections.
HEK-293 and SW480 cells were cultured at 37°C and 7.5% CO2 in DMEM supplemented with 10% FCS, penicillin (50 units/ml), streptomycin (50 μg/ml), and kanamycin (100 μg/ml). Expression constructs of hemagglutinin-tagged MED1 were generated in CMV5 by replacing the Akt gene (15) with the MED1 cDNA. Cells were transfected by using Lipofectamine (Life Technologies, Gaithersburg, MD) and lysed as described previously (16). Three different lysis buffers were used, containing 0.5% NP-40 (15), 0.2% NP-40 (17), or 1% Triton X-100 (18).
Immunoprecipitation and Western Blotting.
Immunoprecipitations were carried out as described previously (16), except that lysates were incubated with antihemagglutinin tag HA.11 beads (Babco, Richmond, CA) for 16 hr. Immune complexes were resolved by 8.5% SDS/PAGE and transferred to Immobilon P membranes (Millipore). Membranes were probed with anti-MLH1 mAb (dilution, 1:250) (PharMingen) and HA.11 antibody (dilution, 1:1,000) (Babco). Detection was carried out by using enhanced chemiluminescence (ECL, Amersham).
Production and Purification of Recombinant Proteins.
PCR-generated fragments corresponding to the entire MED1 ORF or to isolated domains were cloned in pET28(b) (Novagen) and propagated in XL-1 Blue. Constructs were sequenced to verify that unwanted mutations were not inadvertently introduced, and they were transferred into BL21(DE3)(pLysS). These cells were grown to OD600 of 0.4 and induced with 1 mM isopropyl β-d-thiogalactoside at 30 or 37°C for 2 hr. His-tagged proteins were purified from bacterial lysates over a nickel-agarose column (Qiagen), followed by ion-exchange chromatography over SP-Sepharose (Pharmacia). Purity of the proteins was estimated at 95–98% by SDS/PAGE and Coomassie blue staining.
Electrophoretic Mobility-Shift Assay (EMSA).
For EMSA, the methylated probe was assembled by annealing the two complementary oligonucleotides: 5′-AATCCTAMGTGACAMGATGTGMGCAATGMGATGACT-3′ and 5′-AGTCATMGCATTGMGCACATMGTGTCAMGTAGGATT-3′ (M = 5-methylcytosine). The unmethylated and hemimethylated probes were assembled with two complementary oligonucleotides of identical sequence except that cytosine replaced 5-methylcytosine on both strands or only the lower strand, respectively. End-labeling was conducted with [γ-32P]ATP (7,000 Ci/mmol; ICN) and T4 polynucleotide kinase (New England Biolabs). DNA-binding reactions were carried out in 20 mM Hepes, pH 7.9/3 mM MgCl2/10% glycerol/0.1%Triton X-100/0.5 mM EDTA/0.5 mM DTT and contained, as nonspecific competitor DNA, 0.15 μg of poly(dA)/poly(dT) (ICN) and 10 ng of double-strand DNA generated by annealing the two complementary oligonucleotides: 5′-AATCCTAGCTGACAGCATGTGGCCAATGGCATGACT-3′ and 5′-AGTCATGCCATTGGCCACATGCTGTCAGCTAGGATT-3′. Purified methyl-CpG-binding domain (MBD) (20 ng) was incubated with 32P-labeled oligonucleotides (40,000 cpm, 0.1 ng) on ice for 30 min. For competition, MBD was preincubated on ice for 20 min with a 100-fold excess of cold oligonucleotide (10 ng). Binding reactions were loaded on a 10% acrylamide gel and run at 4°C in 0.5× TBE (45 mM Tris/45 mM boric acid/1 mM EDTA, pH 8.3). Dried gels were exposed to autoradiography.
Endonuclease Assay.
Recombinant wild-type MED1 and deletion mutants were incubated with 0.25 μg of the 3.9-kb supercoiled plasmid pCR2 (Invitrogen) at 37°C for 30 min in a buffer containing 20 mM Tris⋅HCl, pH 7.5/150 mM KCl/10 mM MgCl2/1 mM DTT/0.1 mg/ml BSA. Reaction products were separated on a 1% agarose gel buffered in 1× TAE (40 mM tris·acetate/1 mM EDTA, pH 8.0) and containing 0.25 μg/ml of ethidium bromide. For kinetic analysis, approximately 170 fmol of protein was incubated with 1.5 μg of pCR2 plasmid at 37°C. Aliquots of the reactions were removed at appropriate time points and incubated at 65°C in the presence of 0.1% SDS and 10 mM EDTA for 30 min before electrophoresis. Densitometric analysis was performed with nih image (http://rsb.info.nih.gov/nih-image/).
Mutagenicity Assay.
SW480 cl.1 cells were transfected with hemagglutinin-tagged wild-type MED1 and deletion mutants, constructed in pcDNA3.1+ (Invitrogen). Forty-eight hours after transfection, cells were selected with G418 (Life Technologies) at 400 μg/ml for 2 weeks. After confirming expression of the transgenes by Western blot, pooled cultures, maintained in 200 μg/ml G418, were retransfected with the pCAR-OF and pCAR-IF plasmids (19). Cells then were selected with hygromycin (Sigma) for 3 weeks at 300 μg/ml. For β-galactosidase (β-gal) staining, pooled cultures, grown to confluency, were fixed with 10% formalin in PBS for 5 min and stained with 0.8 mg/ml 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) (Sigma) in PBS, in the presence of 5 mM potassium ferrocyanide/5 mM potassium ferricyanide/2 mM MgCl2. For image analysis, randomly selected images of optical fields were acquired with a charge-coupled device camera (Optronics International, Chelmsford, MA) attached to a Nikon invertoscope. Image files were analyzed with the Q500MC Image analysis system (Leica).
RESULTS
Because MMR proteins are known to function in macromolecular complexes, we reasoned that an approach toward the cloning of novel eukaryotic MMR genes would be to identify proteins that interact with known MMR components. In particular, by analogy to the MutL–MutH interaction in the bacterial system, a eukaryotic MMR endonuclease would be expected to interact with the MutL homologue MLH1. Accordingly, we employed the yeast interaction trap, a cloning strategy that screens for protein–protein interactions in S. cerevisiae (13).
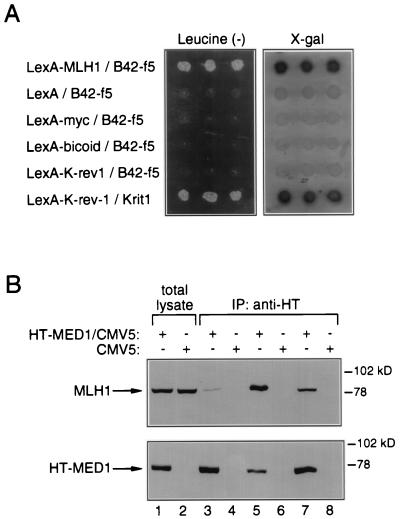
We used human MLH1 (20, 21) fused to the DNA-binding domain of LexA as bait and a human fetal brain cDNA library fused to the B42 activation domain as prey. Twenty-two Leu (+)/lacZ (+) clones (f1–f22) expressing putative MLH1 interactors were selected. Clone f5, which interacted strongly with MLH1, based on the early appearance of colonies on Leu (−)/galactose plates and on the intensity of color formation by colonies grown on X-Gal/galactose plates, was selected for further study. The specificity of the f5-MLH1 interaction was verified by supertransforming virgin EGY191 cells with lacZ reporter, f5 plasmid DNA, and pEG202-t-MLH1 or irrelevant bait constructs. Cells transformed with the combination of f5 and MLH1 bait, but not control cells, grew on Leu (−)/galactose and turned blue on X-Gal/galactose plates (Fig. 1A).
Figure 1.
Interaction between f5/MED1 and MLH1. (A) Specific association of f5 and MLH1 by yeast interaction trap. EGY191 cells were cotransformed with various combinations of plasmids, as indicated, along with the lacZ reporter pSH18–34. Individual transformants were replated onto Leu (−)/galactose plates to score for activation of the LEU2 reporter (Left) and onto X-Gal/galactose plates to score for activation of the lacZ reporter (Right). All interactions were galactose-specific. K-rev-1 and Krit1 represent a positive control for interaction. (B) Coimmunoprecipitation of MED1 and MLH1 from human cells. A band reacting with the anti-MLH1 antibody and comigrating with MLH1 is detected by Western blotting in antihemagglutinin immunoprecipitates from HT-MED1/CMV5-transfected HEK-293 cells and not from CMV5-transfected control cells (Upper). Western blotting of a parallel gel with the antihemagglutinin antibody confirms expression of the HT-MED1 construct in transfected HEK-293 cells (Lower). Lysis buffers contained 0.5% Nonidet P-40 (lanes 1–4), 0.2% Nonidet P-40 (lanes 5 and 6), or 1% Triton X-100 (lanes 7 and 8).
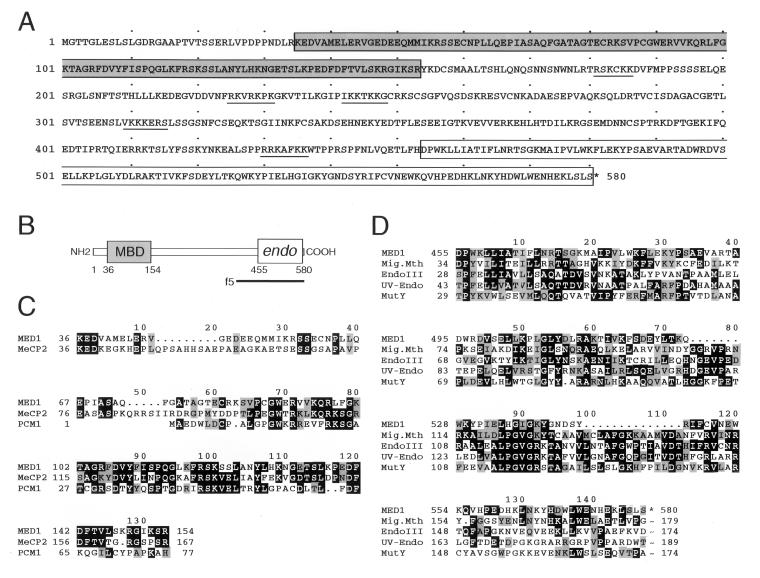
A Northern blot containing mRNA from multiple human tissues probed with the f5 insert revealed that this gene is expressed as a transcript of approximately 2.4 kb (data not shown). To extend the ORF of f5 at the 5′ end, an f5-derived probe was used to screen four additional human cDNA libraries, and 17 clones were isolated. Analysis of these clones allowed the assembly of a nearly complete (2.1-kb) sequence of the gene, which contains an ORF of 1,740 bases, predicting a protein of 580 aa (Fig. 2A).
Figure 2.
Sequence and domain organization of MED1. Predicted amino acid sequence (A) and schematic structure (B) of human MED1; the MBD and the endonuclease domain (endo) are enclosed in the gray and white box, respectively. Putative nuclear localization signals are underlined. The bar depicts the sequence of the original f5 clone. (C) Homology of the MBD region of MED1 to the MBD of the rat protein MeCP2 and the human protein PCM1. (D) Comparison of the endonuclease region of MED1 with related sequences from M. thermoautotrophicum mismatch glycosylase (Mig.Mth), E. coli endonuclease III (EndoIII), M. luteus UV endonuclease (UV-Endo), and E. coli MutY. Identical amino acids are boxed; similar amino acids are shaded.
Analysis of the predicted protein sequence reveals a tripartite architecture that includes known functional domains (Fig. 2B). Near the amino terminus, the f5 protein contains a region of homology with the MBD of MeCP2 and PCM1 (Fig. 2C), two chromosomal proteins that bind CpG-methylated DNA and are involved in transcriptional repression and chromatin stabilization (22, 23). The central portion lacks a recognizable domain, but contains putative nuclear localization signals (Fig. 2A). At the carboxyl terminus, f5 contains a putative catalytic domain with homology to several bacterial, damage-specific endonucleases with glycosylase/lyase activity involved in DNA repair (Fig. 2D), including MutY (24) and endonuclease III (25) from E. coli, mismatch glycosylase Mig.Mth from Methanobacterium (26), and UV-repair endonuclease from Micrococcus luteus (27). The carboxyl-terminal portion of the central region and the catalytic domain (amino acids 406–580) are sufficient for interaction with MLH1, because these sequences are contained in the original f5 clone (Fig. 2B). Because of its domain organization and biochemical activities (see below), f5 was named MED1, for methyl-CpG binding endonuclease 1.
To confirm that the MLH1/MED1 interaction detected in yeast also occurs in human cells, coimmunoprecipitation experiments were performed. HEK-293 cells were transfected with hemagglutinin-tagged MED1 (HT-MED1) or with empty vector. Cell lysates were prepared and immunoprecipitations were carried out with antihemagglutinin tag antibody. Immunoprecipitated proteins separated by SDS/PAGE were transferred to a membrane and probed with an anti-MLH1 antibody. The antibody detected a band of approximately 82 kDa comigrating with MLH1 in the antihemagglutinin immunoprecipitate from HT-MED1-transfected HEK-293 cells but not from control cells (Fig. 1B). This experiment suggests that MED1 can form a complex with MLH1 in vivo.
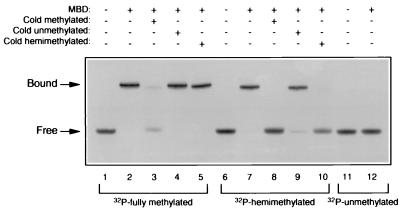
The DNA-binding properties of MED1 were analyzed by EMSA. To avoid endonucleolytic degradation of probe and competitor DNA by full-length MED1, EMSA analysis was conducted with the MED1 MBD (codons 1–154). Purified MBD was incubated with a 32P-labeled double-strand oligonucleotide of arbitrary sequence containing four symmetrical methyl-CpG sites (fully methylated oligo). MBD also was incubated with 32P-labeled double-strand oligonucleotides of identical sequence in which cytosines replaced methyl-cytosines on one or both strands (hemimethylated and unmethylated oligos). EMSA analysis indicated that the MED1 MBD binds to fully and hemimethylated DNA and fails to bind to unmethylated DNA (Fig. 3, lanes 2, 7, and 12). Binding to the fully methylated probe was competed by preincubation with a 100-fold excess of cold, fully methylated oligonucleotide (Fig. 3, lane 3), whereas no competition was observed with the hemimethylated or unmethylated oligonucleotides (Fig. 3, lanes 4 and 5). Binding to the hemimethylated probe was competed by preincubation with the fully or hemimethylated oligonucleotides (Fig. 3, lanes 8 and 10); little competition was observed with the unmethylated oligonucleotide (Fig. 3, lane 9). This experiment supports the methyl-CpG-binding specificity of the MED1 MBD and suggests that the binding affinity of MED1 to fully methylated DNA is higher than that to hemimethylated DNA.
Figure 3.
Mobility-shift assay of MED1 MBD with fully methylated, hemimethylated, and unmethylated DNA probes. The MED1 MBD binds to 32P-labeled double-stranded oligonucleotides containing four fully methylated (lane 2) or hemimethylated CpG sites (lane 7). No binding is detected when the unmethylated probe is used (lane 12). For binding competition, the cold oligonucleotides used are indicated.
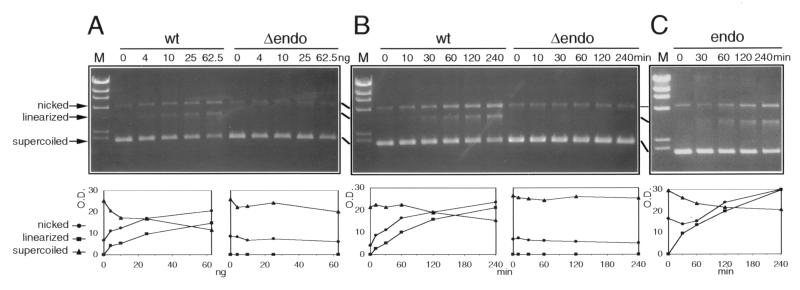
MED1 endonuclease activity was assayed by evaluating the conversion of supercoiled plasmid DNA into nicked and linear molecules. Incubation of supercoiled DNA with wild-type MED1 resulted in a dose-dependent appearance of nicked and linearized molecules (Fig. 4A). Similarly, incubation of a constant amount of MED1 with the supercoiled plasmid generated nicked and linearized molecules with approximately similar linear kinetics (Fig. 4B). To rule out the possibility that a bacterial endonuclease activity copurifying with MED1 is responsible for the observed effects, we also purified a deletion mutant lacking the putative endonuclease domain (codons 1–454). This mutant failed to produce nicked and linearized DNA molecules (Fig. 4 A and B). A similar experiment with the isolated endonuclease domain (codons 455–580) also resulted in the appearance of nicked and linearized DNA molecules (Fig. 4C), demonstrating that this domain is sufficient for MED1 single- and double-strand endonuclease activity. Digestion of the MED1-linearized plasmid with the restriction enzyme EcoRI, which performs two closely spaced cuts on this plasmid, resulted in the appearance of a smear, indicating that MED1 does not have preferential cutting sites on this substrate (data not shown).
Figure 4.
Endonuclease activity of MED1. (A) The indicated increasing amounts of wild-type MED1 (wt) and a deletion mutant lacking the endonuclease domain (Δendo) were incubated with 0.25 μg of supercoiled pCR2 plasmid at 37°C for 30 min. Reaction products were analyzed by agarose gel electrophoresis (Upper). The migration of nicked, linearized, and supercoiled DNA is indicated. A small amount of nicked molecules is present in the plasmid DNA substrate at time zero. M, λ/HindIII digest size standards. (Lower) Densitometric analysis of the nicked (circles), linearized (squares), and supercoiled (triangles) bands. OD, in thousands of units. (B) Time course of MED1 endonuclease activity. MED1 wt and Δendo (12.5 ng, approximately 170 fmol) were incubated with 1.5 μg (approximately 580 fmol) of pCR2 plasmid at 37°C; at the indicated time points, an aliquot of the reaction was removed for electrophoretic analysis. (C) Endonuclease activity of the recombinant MED1 endonuclease domain (codons 455–580, endo). Endo (3.6 ng, approximately 170 fmol) was incubated with 1.5 μg (approximately 580 fmol) of pCR2 plasmid at 37°C and processed as in B.
To assess the physiological significance of MED1 in DNA repair, we conducted cell transfection studies. The modular structure of MED1, with an amino-terminal domain involved in binding to methylated DNA and a carboxyl-terminal region involved in catalysis and complex formation with MLH1, suggests that deletion of one domain might generate mutants with dominant-negative properties. Because the endonuclease domain is sufficient for catalytic activity (Fig. 4C), the MBD domain may be required for localization of MED1 to DNA regions undergoing repair in vivo. If this were the case, the ΔMBD mutant would be expected to form nonfunctional, mislocalized complexes with MLH1 and possibly other components of the repair system and, thus, alter MMR.
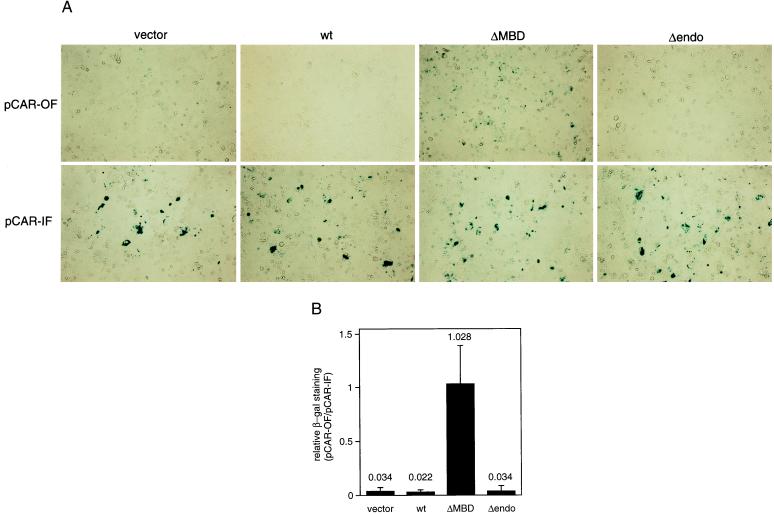
A clone of SW480 colon carcinoma cells, named SW480 cl.1, was isolated by limiting dilution and, before transfection, analyzed for MSI to verify that it retained the MMR proficiency of parental SW480 cells. SW480 cl.1 cells were transfected with empty vector or expression constructs of hemagglutinin-tagged wild-type MED1 and two deletion mutants, ΔMBD (codons 155–580) and Δendo (codons 1–454). Stable cell lines (pooled cultures) were generated and they were retransfected with the episomal vector pCAR-OF containing an out-of-frame (CA)28 repeat within the coding region of the β-gal gene. Insertions/deletions in the (CA) repeat may reconstitute the reading frame and restore β-gal activity of this reporter, suggesting a defect in MMR activity (19). This assay has been employed to demonstrate the dominant-negative phenotype of a PMS2 deletion mutant (19). As a control for the efficiency of transfection/expression, cell lines also were transfected with the pCAR-IF vector containing the β-gal gene with an in-frame (CA)27 repeat (19). After selection of episome-containing cells, pooled cultures were stained with X-gal to assess β-gal activity. Only few blue cells per optical field were detected in wild-type MED1-, Δendo-, or control vector-transfected cells, whereas a large number of blue cells per optical field was present in cells transfected with the ΔMBD mutant (Fig. 5A). To estimate the relative amount of blue cells, 10 randomly selected images of optical fields from each culture were captured with a charge-coupled device camera and digitally analyzed. Relative β-gal staining for vector-, wild-type MED1-, ΔMBD-, and Δendo-containing cell lines was expressed as the ratio between the percentage of blue area in optical fields of pCAR-OF- and pCAR-IF-transfected cultures. This adjustment normalizes for differences in transfection/expression efficiency among the various cell lines (19). The results indicate that ΔMBD-transfected cultures contain approximately 30-fold more blue cells than other cultures (Fig. 5B). To rule out tissue culture artifacts, the entire experiment (transfection, selection, and staining) was conducted in triplicate and yielded reproducible results.
Figure 5.
(A) In situ β-gal staining of pooled cultures of SW480 cl.1 cells stably transfected with empty vector, wild-type MED1 (wt), or deletion mutants (ΔMBD and Δendo) and subsequently transfected with β-gal reporter genes containing an out-of-frame (pCAR-OF, Upper) or in-frame (pCAR-IF, Lower) (CA) microsatellite repeat. Representative fields of a triplicate experiment are shown. β-gal staining is approximately equal in the pCAR-IF-transfected cell lines expressing different MED1 constructs, whereas, among the pCAR-OF-transfected cell lines, only the ΔMBD-expressing culture contains many blue cells. (B) Relative β-gal staining of mass cultures of SW480 cells stably transfected with empty vector, wild-type MED1 (wt), or deletion mutants (ΔMBD and Δendo). Relative β-gal staining represents the ratio between the percentage of blue area in optical fields of pCAR-OF- and pCAR-IF-transfected cultures. Data are the mean ± SD derived from the analysis of 10 optical fields for each culture.
Although we cannot formally rule out the possibility that the hypermutability associated with the ΔMBD mutant is the result of an enhanced, deregulated endonuclease activity, this seems to be unlikely. In fact, this mutant and the endo mutant (that also lacks the MBD domain) have in vitro catalytic activity comparable to wild-type protein (Fig. 4 B and C, and data not shown). This suggests that the ΔMBD mutant acts in a dominant-negative fashion.
DISCUSSION
A long-standing issue in eukaryotic MMR is defining the nature of the strand-discrimination process, i.e., the mechanism(s) responsible for targeting repair to the newly synthesized DNA. In E. coli the single-strand endonuclease MutH performs this function via interaction with MutL and recognition of hemimethylated d(GATC) sites. In an effort to identify novel human MMR genes that may help to elucidate this problem, we performed a yeast interaction trap screening by using MLH1 as bait, and we cloned MED1. Although the MED1 protein does not display a significant sequence homology to MutH, it appears to be a viable candidate for a eukaryotic functional homologue of this bacterial enzyme. MED1 has several features compatible with such a role: first, it interacts with MLH1; second, it has homology to bacterial DNA repair enzymes and displays endonuclease activity; third, it binds with different affinity to, hence discriminates between, fully methylated and hemimethylated CpG sequences; and, fourth, it appears to be involved in stability of microsatellite sequences.
Because we have functionally assayed microsatellite instability, which is not necessarily linked to a MMR defect, we cannot definitely conclude that MED1 is an MMR protein. Stability of microsatellite sequences may be affected by DNA replication and recombination (28); so, it is also possible that MED1 is involved in these DNA processes. Demonstration that MED1 is a bona fide MMR protein will have to await elucidation of its potential role in repair of mismatch-containing substrates in in vivo or in vitro reconstituted systems. In particular, if MED1 is a MMR protein, it will be important to determine whether, like MutH, it is involved in strand discrimination. In addition, because E. coli MutL is involved in repair pathways other than the long-patch, methyl-directed system, such as the Vsr-dependent short-patch MMR pathway (29), it will be interesting to elucidate whether MED1 participates in similar reactions. Finally, based on its homology to bacterial damage-specific glycosylases/lyases, the possibility that MED1 functions in a pathway of base excision repair should be taken into consideration.
The low endonuclease activity of MED1 on supercoiled DNA substrate may be due to the absence of posttranslational modifications in the recombinant protein. Alternatively, it may reflect the lack of critical factors present in vivo, such as hMutSα (30), hMutSβ (31), and hMutLα (32). Likewise, E. coli MutH has a weak Mg2+-dependent endonuclease activity, which is augmented 30-fold when MutS, MutL, and ATP are present (33). In addition, it will be important to determine whether MED1 activity is modulated by binding to CpG methylated DNA; given the ability of MED1 to discriminate between fully and hemimethylated DNA, the DNA methylation state may be relevant to its catalytic properties.
The domain structure and biochemical properties of MED1 imply a potential role for cytosine methylation in eukaryotic DNA repair, perhaps MMR. Although the transfection experiments with the ΔMBD mutant of MED1 indicate that the MBD domain is important for MED1 function, the precise role of recognition of CpG-methylated DNA needs to be established. An attractive hypothesis is that, similar to the bacterial methyl-directed reaction, strand specificity in human MMR could be determined, at least in part, by the MED1-mediated recognition of transiently hemimethylated CpG sites generated after DNA replication. DNA termini generated at the replication fork may represent the only strand-targeting signal in DNA methylation-deficient eukaryotes, such as Drosophila, Caenorhabditis elegans, and S. cerevisiae (3, 4). However, in monkey CV1 cells, CpG hemimethylation was shown to synergize with single-strand nicks in directing repair to the unmethylated strand (34). Our findings are consistent with recent studies indicating that DNA methylation plays a role in maintaining genomic stability (35, 36). Because lower eukaryotes lack genome-wide methylation, this role is likely to be adjunct or auxiliary, but, nevertheless, it may be very important for genomic fidelity in mammals.
Based on the preceding data suggesting a possible role in MMR, the MED1 gene could be involved in the pathogenesis of hereditary nonpolyposis colorectal cancer (HNPCC) and sporadic colorectal cancers with MSI. Because only 50–70% of HNPCC and sporadic colorectal tumors with MSI carry mutations in the known MMR genes (5–8), MED1 mutations may account for some of the remaining cases.
Future biochemical and genetic experiments will address the hypothesis that MED1 activity is regulated by methyl-cytosine binding and interaction with MMR components and will examine whether MED1 mutations are relevant to the pathogenesis of human cancer.
Acknowledgments
We would like to thank A. Knudson for constant advice; J. Chernoff, J. Feng, R. Katz, A. Knudson, E. Moss, R. Perry, A. M. Skalka, and J. R. Testa for critical reading of the manuscript; B. Vogelstein for the gift of the pCAR-OF and pCAR-IF plasmids; and A. K. Godwin and G. Kruh for the gift of the C200 cDNA library. A.B. thanks his wife, his family, Mrs. E. Cavallaro Scarlato, P. Engstrom, and P. Tsichlis for continuous support and encouragement. This work was supported by American Cancer Society Grant IRG-191, Public Health Service Grants CA78412-01 and CA06927, Italian Association for Cancer Research Special Project “Hereditary Colorectal Tumors,” and Italian National Research Council Contract 97.04207.CT04. Additional support was provided by an appropriation from the Commonwealth of Pennsylvania to the Fox Chase Cancer Center.
ABBREVIATIONS
- MMR
DNA mismatch repair
- MBD
methyl-CpG-binding domain
- MSI
microsatellite instability
- EMSA
electrophoretic mobility-shift assay
- β-gal
β-galactosidase
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
Note Added in Proof
After submission of this manuscript, it was brought to our attention that MED1 is identical to MBD4, a recently described protein that binds to methylated DNA (37).
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF114784).
References
- 1.Modrich P. Science. 1994;266:1959–1960. doi: 10.1126/science.7801122. [DOI] [PubMed] [Google Scholar]
- 2.Fishel R, Kolodner R D. Curr Opin Genet Dev. 1995;5:382–395. doi: 10.1016/0959-437x(95)80055-7. [DOI] [PubMed] [Google Scholar]
- 3.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 4.Jiricny J. EMBO J. 1998;17:6427–6436. doi: 10.1093/emboj/17.22.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch H T, Smyrk T. Cancer. 1996;78:1149–1167. doi: 10.1002/(SICI)1097-0142(19960915)78:6<1149::AID-CNCR1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama Y, Sato H, Yamada T, Nagasaki H, Tsuchiya A, Abe R, Yuasa Y. Cancer Res. 1997;57:3920–3923. [PubMed] [Google Scholar]
- 8.Liu B, Nicolaides N C, Markowitz S, Willson J K V, Parsons R E, Jen J, Papadopolous N, Peltomäki P, de la Chapelle A, Hamilton S R, et al. Nat Genet. 1995;9:48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 9.Perucho M. Biol Chem. 1996;377:675–684. [PubMed] [Google Scholar]
- 10.Holmes J, Clark S, Modrich P. Proc Natl Acad Sci USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas D C, Roberts J D, Kunkel T A. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- 12.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 13.Golemis E, Gyuris J, Brent R. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D, Seidman J G, Smith J, Struhl K, editors. New York: Wiley; 1996. pp. 20.1.1–20.1.28. [Google Scholar]
- 14.Bellacosa A, Datta K, Bear S E, Patriotis C, Lazo P A, Copeland N G, Jenkins N A, Tsichlis P N. J Virol. 1994;68:2320–2330. doi: 10.1128/jvi.68.4.2320-2330.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datta K, Franke T F, Chan T O, Makris A, Yang S I, Kaplan D R, Morrison D K, Golemis E A, Tsichlis P N. Mol Cell Biol. 1995;15:2304–2310. doi: 10.1128/mcb.15.4.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellacosa A, Chan T O, Ahmed N N, Datta K, Malstrom S, Stokoe D, McCormick F, Feng J, Tsichlis P N. Oncogene. 1998;17:313–325. doi: 10.1038/sj.onc.1201947. [DOI] [PubMed] [Google Scholar]
- 17.Gu W, Bhatia K, Magrath I T, Dang C V, Dalla-Favera R. Science. 1994;264:251–254. doi: 10.1126/science.8146655. [DOI] [PubMed] [Google Scholar]
- 18.Law S F, Estojak J, Wang B, Mysliwiec T, Kruh G, Golemis E A. Mol Cell Biol. 1996;16:3327–3337. doi: 10.1128/mcb.16.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolaides N C, Littman S J, Modrich P, Kinzler K W, Vogelstein B. Mol Cell Biol. 1998;18:1635–1641. doi: 10.1128/mcb.18.3.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bronner C E, Baker S M, Morrison P T, Warren G, Smith L G, Lescoe M K, Kane M, Earabino C, Lipford J, Lindblom A, et al. Nature (London) 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 21.Papadopoulos N, Nicolaides N C, Wei Y F, Ruben S M, Carter K C, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, et al. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 22.Nan X, Campoy F J, Bird A. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 23.Cross S H, Meehan R R, Nan X, Bird A. Nat Genet. 1997;16:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- 24.Michaels M L, Pham L, Nghiem Y, Cruz C, Miller J H. Nucleic Acids Res. 1990;18:3841–3845. doi: 10.1093/nar/18.13.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asahara H, Wistort P M, Bank J F, Bakerian R H, Cunningham R P. Biochemistry. 1989;28:4444–4449. doi: 10.1021/bi00436a048. [DOI] [PubMed] [Google Scholar]
- 26.Horst J P, Fritz H J. EMBO J. 1996;15:5459–5469. [PMC free article] [PubMed] [Google Scholar]
- 27.Piersen C E, Prince M A, Augustine M L, Dodson M L, Lloyd R S. J Biol Chem. 1995;270:23475–23484. doi: 10.1074/jbc.270.40.23475. [DOI] [PubMed] [Google Scholar]
- 28.Kunkel T A. Nature (London) 1993;365:207–208. doi: 10.1038/365207a0. [DOI] [PubMed] [Google Scholar]
- 29.Lieb M, Bhagwat A S. Mol Microbiol. 1996;20:467–473. doi: 10.1046/j.1365-2958.1996.5291066.x. [DOI] [PubMed] [Google Scholar]
- 30.Drummond J T, Li G M, Longley M J, Modrich P. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 31.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 32.Li G M, Modrich P. Proc Natl Acad Sci USA. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Au K G, Welsh K, Modrich P. J Biol Chem. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 34.Hare J T, Taylor J H. Proc Natl Acad Sci USA. 1985;82:7350–7354. doi: 10.1073/pnas.82.21.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lengauer C, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1997;94:2545–2550. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen R Z, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. Nature (London) 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 37.Hendrich B, Bird A. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]