Abstract
Expression of the DMP1 transcription factor, a cyclin D-binding Myb-like protein, induces growth arrest in mouse embryo fibroblast strains but is devoid of antiproliferative activity in primary diploid fibroblasts that lack the ARF tumor suppressor gene. DMP1 binds to a single canonical recognition site in the ARF promoter to activate gene expression, and in turn, p19ARF synthesis causes p53-dependent cell cycle arrest. Unlike genes such as Myc, adenovirus E1A, and E2F-1, which, when overexpressed, activate the ARF-p53 pathway and trigger apoptosis, DMP1, like ARF itself, does not induce programmed cell death. Therefore, apart from its recently recognized role in protecting cells from potentially oncogenic signals, ARF can be induced in response to antiproliferative stimuli that do not obligatorily lead to apoptosis.
Keywords: p53, cyclin D
The singularly most frequently disrupted gene in cancer is p53, whose loss of function occurs in more than half of human tumors (1). The p53 protein serves as an integrator of different cellular stress responses initiated by DNA damage, hypoxia, and hyperproliferative oncogenic signals (2, 3). In its role as a transcription factor, it activates a series of genes that can restrict cell cycle progression and trigger apoptosis. Among p53’s known transcriptional targets is Mdm2, which acts in a feedback loop to antagonize p53 function (4, 5). Mdm2 binding inhibits p53 transcriptional activity (6, 7), induces p53 ubiquitination (8–10), and accelerates p53 nuclear export and its destruction in cytoplasmic proteasomes (11).
INK4a/ARF is perhaps the second most commonly disrupted locus in cancer cells (12). It encodes two distinct tumor suppressor proteins: p16INK4a, which inhibits the phosphorylation of the retinoblastoma protein by cyclin D-dependent kinases (13), and p19ARF (14), which stabilizes and activates p53 to promote either cell cycle arrest or apoptosis (reviewed in ref. 15). ARF acts to check potentially harmful growth promoting signals conveyed by overexpression of c-Myc, E2F-1, adenovirus E1A, or the Abelson oncogene (v-Abl) (16–19), but it is not required for p53 activation in response to DNA damage by radiation or genotoxic drugs (20, 21). The p19ARF protein binds directly to Mdm2 to neutralize its functions, thereby potentiating p53 transcriptional activity (21–24). Hence, loss of ARF limits cell-autonomous tumor surveillance in response to particular oncogenic signals, and animals lacking ARF function, such as those lacking p53, are highly tumor prone (20). Not surprisingly, human cancer cells that retain p53 function overexpress Mdm2 (25) or sustain deletions that dismantle ARF function (12).
In attempting to determine how ARF is regulated, we noted that the mouse ARF promoter contains a potential binding site for a recently discovered transcription factor designated DMP1 (26). DMP1 was isolated in a yeast two-hybrid interactive screen performed with cyclin D2 as bait, and the protein binds to any of the three D-type cyclins, but not to cyclins A, B, C, or H in vitro or when expressed with them in insect Sf9 or mammalian cells (26, 27). DMP1 is a 761-amino acid protein that contains a central DNA-binding domain composed of three imperfect Myb-like repeats flanked by acidic activating domains at both its N and C termini. The cognate human and mouse proteins are 95% identical, and hDMP1 on human chromosome 7q21 is frequently deleted in myeloid leukemia, connoting a possible role for DMP1 as a tumor suppressor (28). DMP1 binds to nonameric consensus DNA sequences containing G-G/T-A cores; those that contain GGA can also be bound by certain Ets family transcription factors (26). D-type cyclins associate with a domain in DMP1 located just N-terminal to the Myb repeats, thereby antagonizing the ability of DMP1 to bind DNA and to activate gene expression (27). Interestingly, these interactions do not depend on the D-type cyclin-dependent kinases, CDK4 and CDK6. In fact, CDK4 and DMP1 form mutually independent complexes with D-type cyclins, and inhibitors of CDK4 do not abrogate interactions between these cyclins and DMP1. It should be noted that CDK-independent functional interactions between D-type cyclins and transcription factors are not unprecedented and have been observed with the estrogen receptor (29, 30) and with other Myb family members (31).
DMP1 is ubiquitously expressed at low levels in mouse cell lines and tissues, but is more prominent in nondividing cells and may facilitate cell differentiation in certain lineages (26, 27, 32). Importantly, enforced expression of DMP1 in mouse fibroblasts can induce cell cycle arrest (27). This led us to consider the possibility that genes encoding negative regulators of cell cycle progression might be direct targets of DMP1 regulation. Here we show that DMP1 activates the murine ARF promoter and induces cell cycle arrest in primary diploid mouse fibroblasts in an ARF-dependent manner.
MATERIALS AND METHODS
Cell Culture.
Primary mouse embryonic fibroblasts (MEFs) explanted at E13.5–14.5 of gestation were maintained in DMEM plus 10% fetal bovine serum, 2 mM glutamine, 0.1 mM nonessential amino acids, 55 μM 2-mercaptoethanol, and 10 μg/ml gentamicin (20). NIH 3T3 and Balb-3T3 (10–1) cells were cultured in DMEM plus 10% fetal bovine serum, 2 mM glutamine, with 100 units/ml each of penicillin and streptomycin.
Cloning of Murine ARF Promoter.
A 129/SvjE mouse genomic library was screened with an ARF-specific cDNA probe (14). A 5.0-kb EcoRI fragment isolated from phage was subcloned into pBluescript, and a 990-bp SmaI fragment hybridizing to the probe was subcloned and sequenced. A 281-bp BamHI–BglII DNA subdomain containing a minimal promoter region was ligated to a luciferase reporter gene to yield plasmid pGL2-ARFpro BamHI. To mutate the single DMP1 consensus site in the promoter, the plasmid was digested with KpnI and ApaI and ligated with mutant oligonucleotides obtained by annealing 5′-CGGATCCGGAGCGTGCCCTGCGCGGGAGGCAGCGGGACCCCGTCGACGGCAGGGCC-3′ (sense) and 5′-CTGCCGTCGACGGGGTCCCGCTGCCTCCCGCGCAGGGCACGCTCCGGATCCGGTAC-3′ (antisense) with mutated nucleotides in both strands underlined.
Virus Production and Infection.
Human kidney 293T cells were transfected with a helper ecotropic retrovirus plasmid defective in psi-2 packaging sequences, together with pSRα vectors containing murine DMP1, human c-Myc, human E2F-1, or human Ets1 cDNAs (16, 32). Viruses were harvested every 6 hr 24–72 hr after transfection, filtered, and stored at 4°C until used for infection (16). To construct a retroviral vector containing DMP1 linked to a mutated tamoxifen-responsive element of the estrogen receptor (DMP1-ER), a 3′ 0.3-kb EcoNI DNA fragment of murine DMP1 cDNA was amplified by PCR using 5′-CACTGACCTTAAGCAGGAAG-3′ (sense) and 5′-AGAAGCTTGGATCCGTGTGACAGTTTACTAAGTCCTC-3′ (antisense) primers (HindIII site italicized and BamHI site underlined). This removed the translational stop codon and allowed insertion of DMP1 sequences 5′ and in frame to those encoding the ER element. The product was digested with EcoNI and HindIII and used to replace the cognate 3′ DMP1 cDNA segment in pBluescript. After confirmation of the nucleotide sequence, a 2.4-kb BamHI fragment containing DMP1 coding sequences was cloned into the BamHI site of pBabe-puro retroviral vector containing the ER element (generously provided by Natasha Aziz and Martin McMahon, University of California, San Francisco). Pooled, filtered viruses were used to infect wild-type (passage 3–5) or ARF-null MEFs (2 × 105 cells seeded into 100-mm diameter culture dishes). Cells were infected with three additions of 4 ml of virus-containing supernatant at 5-hr intervals in the presence of 10 μg/ml Polybrene (Sigma). Those infected with DMP1-ER virus were selected 36 hr after infection with 2 μg/ml puromycin for 48 hr before treatment of surviving cells with 1 μM 4-hydroxytamoxifen (4-HT; Sigma).
RNA and Protein Expression.
Quantitative reverse transcription–PCRs employed specific primers for murine ARF exon 1β (30 cycles) and for β-actin (20 cycles) used as a control (33). Protein analyses were performed as described (16, 24). Samples (200 μg of protein per lane) were separated by denaturing electrophoresis and transferred to nitrocellulose membranes (MSI, Westboro, MA) before immunoblotting. Anti-actin (C-11) was from Santa Cruz Biotechnology.
Electrophoretic Mobility-Shift Assay.
Recombinant DMP1 protein was prepared in Sf9 cells (26). Purified bacterial proteins representing the Ets1 DNA-binding domain or full-length Ets1 (32) were originally obtained from Jacques Ghysdael (University of Paris, Orsay, France). Electrophoretic mobility-shift assays were performed (27) using either a 281-bp genomic fragment (−225 to +56) or double-stranded oligonucleotides containing the DMP1/Ets site obtained by annealing oligonucleotide 5′-AATTGGGACCCCGGATGCGGCAG-3′ (sense strand; DMP1/Ets consensus sequence underlined) with a complementary antisense strand. For competition experiments, a 200-fold excess of unlabeled oligonucleotides was added to reaction mixtures before probe. To verify the identity of the proteins in shifted complexes, reaction mixtures were incubated with control nonimmune rabbit serum, serum AF (26), and M-10 (both to DMP1 C-terminal epitopes), S-19 (DMP1 N terminus), or C-20 (Ets1 C terminus) before electrophoresis. M-10, S-19, and C-20 were from Santa Cruz Biotechnology.
Transactivation Assays.
NIH 3T3 cells were transfected with 4 μg of pGL2-ARFpro BamHI or its DMP1-binding site mutant, with or without increasing amount of pFLEX-DMP1, pEVRFO-Ets1, pCMV-Ets2, pRcRSV-Elf1, pdEB-Fli1, pdEB-EWS-Fli1, or pCMV-E2F-1 (89–437), and 4 μg of β-actin secreted alkaline phosphatase (SEAP) control vectors (27, 32, 34, 35). Transfections and normalization of luciferase levels with internal control SEAP levels were performed as described (27).
BrdUrd Incorporation and Immunofluorescence.
Wild-type or ARF-null MEFs (4 × 104 cells) were seeded on gelatin-coated coverslips 16 hr before virus infection. Cells were infected three times with empty vector, or vectors expressing murine DMP1, human c-Myc, human E2F-1, or human Ets1. Cells were labeled 36 hr after virus infection with BrdUrd (Sigma) for 14 hr in complete medium. For pulse labeling, MEFs were treated with 2 μM 4-HT for 36 hr and labeled with BrdUrd for 3 hr at different intervals throughout the inductive phase. Cells fixed in ice-cold methanol/acetone (1:1) for 10 min at −20°C were stained with affinity-purified antibodies to DMP1 (AF) (26), p19ARF (14), c-Myc (Upstate Biotechnology, Lake Placid, NY), or Ets-1 (C-20, Santa Cruz Biotechnology), and counterstained with a 1:1 dilution of mAbs to BrdUrd (Amersham Pharmacia) as described (27).
Apoptosis Assays.
Wild-type MEFs infected with the indicated retroviruses for 36 hr were starved for serum for 24 hr. Viability was determined by trypan blue dye exclusion, and DNA fragmentation was monitored by a terminal deoxynucleotidyltransferase (fluorescence-activated cell sorter/terminal deoxynucleotidyltransferase-dUTP end labeling) assay and by measurement of subdiploid DNA content of propidium iodide-stained nuclei (16).
RESULTS
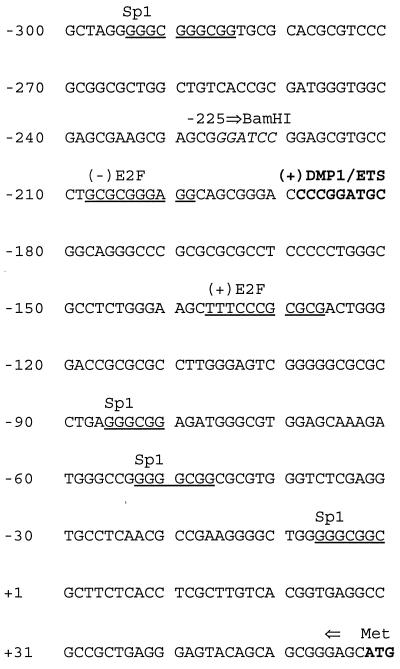
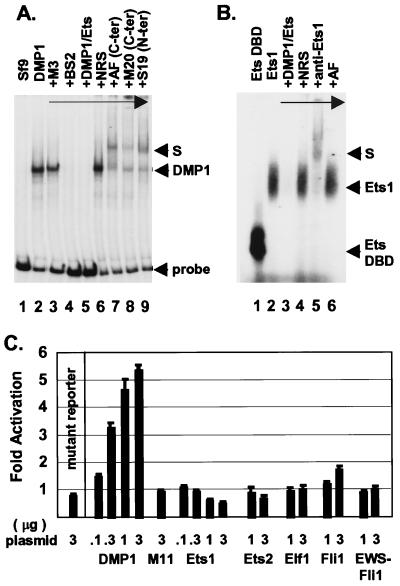
We determined the nucleotide sequence of the proximal promoter region of the murine ARF gene relative to the transcription initiation site as defined by S1 mapping analysis (Fig. 1). Putative binding sites for known transcription factors were identified in a 300-bp region 5′ to the start site, which included a perfect DMP1/Ets consensus at −189 to −181. A recombinant DMP1 protein produced in baculovirus-infected insect Sf9 cells bound to a radiolabeled 281-bp fragment from the ARF promoter encompassing nucleotides −225 to +56 (Fig. 2A, lane 2). Binding of DMP1 to the 281-bp promoter fragment was completely inhibited by a 23-bp oligonucleotide containing the DMP1/Ets consensus sequence (Fig. 2A, lane 5), as well as by a variant oligonucleotide [CCCGTATGT, previously designated BS2 (26)] that lacks the GGA core sequence required for Ets binding (Fig. 2A, lane 4). Conversely, an oligonucleotide containing a reiterated Ets-specific consensus binding site (CCCGGAAGT, designated M3) to which DMP1 cannot bind did not compete (Fig. 2A, lane 3). DNA–protein complexes containing bound DMP1 were further retarded in mobility in the presence of antibodies directed to different DMP1 epitopes (lanes 7–9), but not by nonimmune serum (lane 6).
Figure 1.
Mouse ARF promoter. The nucleotide sequence of 300 bp 5′ to the transcriptional start site (+1) is shown. Putative binding sites for DMP1/ETS (boldface), Sp1, and E2F-1 (both underlined) are indicated; (+) indicates sense and (−) the antisense strand. The translational initiation codon (ATG, boldface) is at +59. The BamHI site (italics) at −225 used to construct a promoter-reporter expression plasmid is indicated by the right arrow.
Figure 2.
DMP1 binds and transactivates the ARF promoter. (A) Electrophoretic mobility-shift assays were performed with a radiolabeled 281-bp ARF promoter fragment (bracketed by arrows in Fig. 1) using recombinant DMP1 made in insect Sf9 cells. Lane 1 shows results with uninfected Sf9 lysates and lane 2 with extracts of cells expressing DMP1. Competition was performed with an Ets-specific (M3, lane 3), DMP1-specific (BS2, lane 4) or a cognate ARF promoter consensus oligonucleotide (lane 5). Nonimmune rabbit serum (lane 6) or different antibodies to DMP1 (lanes 7–9) were added before probe. (B) Electrophoretic mobility-shift assays were performed with a recombinant Ets1 protein (lanes 2–6) or its DNA-binding domain (DBD, lane 1). Competition was performed using the ARF promoter DMP1 consensus site (lane 3). Antibodies to Ets1 (lane 5) or DMP1 (lane 6) were added before probe. (C) Transactivation of the ARF promoter-reporter in NIH 3T3 cells was performed by cotransfection with vectors encoding DMP1, a DMP1 mutant (M11) that cannot bind to DNA, or the indicated Ets family members (abscissa). Plasmid inputs (μg DNA) are indicated at the left. Activation (ordinate) is normalized to SEAP expression.
Because the DMP1 site on the ARF promoter contains a GGA core (Fig. 1), Ets family proteins can also bind to it. A recombinant Ets1 protein produced in bacteria or a segment representing its DNA-binding domain bound to a short 23-bp double-stranded oligonucleotide containing the ARF DMP1 consensus site (Fig. 2B, lanes 1 and 2). Binding was competed by the cognate oligonucleotide (Fig. 2B, lane 3), and the mobility of these complexes was retarded by antiserum to Ets1 but not to DMP1 (lanes 5 and 6).
A plasmid containing the 281-bp ARF promoter fragment ligated to a luciferase reporter gene was transfected into NIH 3T3 fibroblasts. Cotransfection with increasing concentrations of a DMP1 expression vector enhanced reporter gene expression, whereas a DMP1 point mutant (M11) that is unable to bind to DNA had no activity (Fig. 2C). Similarly, DMP1 deletion mutants defective in transactivation (27) were inactive in this assay (data not shown). Mutation of the DMP1 binding site within the ARF promoter (changing CCCGGATGC to CCCGTCGAC) also abolished transactivation by wild-type DMP1 (Fig. 2C, mutant reporter). Therefore, sequences within the −189 to −181 ARF promoter segment were the only ones responsible for DMP1-mediated transactivation. Although Ets1 could bind to a 23-bp oligonucleotide containing the DMP1 consensus site (Fig. 2B), several Ets proteins were unable to induce significant reporter gene expression from the more complex 281-bp ARF promoter (Fig. 2C). Only Fli1 showed minimal activity, whereas Ets1 was slightly inhibitory. Therefore, in the context of the larger promoter fragment, DMP1 binding is strongly preferred.
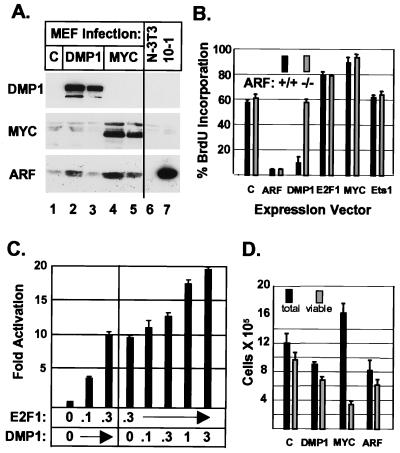
Based on the above observations, we tested whether introduction of DMP1 would induce synthesis of the endogenous ARF protein in normal diploid fibroblast strains. Early-passage MEF strains were infected with retroviral vectors encoding either DMP1 or c-Myc, a known rapid inducer of p19ARF protein expression used here as a positive control (16). In early-passage MEFs, p19ARF levels are low and remained so in cells infected with the naked expression vector (Fig. 3A, lane 1). Both DMP1 (Fig. 3A, lanes 2 and 3) and c-Myc (Fig. 3A, lanes 4 and 5) induced ARF protein synthesis to a similar extent (see Fig. 3 legend for quantitation). In turn, wild-type MEFs infected with DMP1 underwent cell cycle arrest, similar to cells infected with a retrovirus encoding p19ARF itself (Fig. 3B). In direct contrast, MEF strains derived from ARF-null animals were refractory to DMP1-induced arrest, indicating that ARF function was required for inhibition of S phase entry. Cells infected with a vector encoding Ets-1, whether containing or lacking ARF, behaved indistinguishably from those infected with the control vector (Fig. 3B), consistent with the inability of Ets proteins to stimulate reporter gene expression driven by the ARF promoter fragment (Fig. 2C).
Figure 3.
DMP1 induces p19ARF and cell cycle arrest in wild-type but not ARF-null MEFs. (A) Infection of wild-type MEF strains with a DMP1 virus (lanes 2 and 3) or a Myc virus (lanes 4 and 5) induces p19ARF protein. Amounts of protein loaded in lanes 3 and 5 were 40% of those in lanes 2 and 4. All viruses expressed the T cell coreceptor CD8; whereas 95% of Myc-infected cells were CD8 positive (lane 4), only 35% of cells infected with DMP1 virus expressed the CD8 marker (lane 2). NIH 3T3 cells (lane 6) have sustained ARF deletions, whereas 10-1 cells (lane 7) lack p53 and overexpress p19ARF through loss of feedback control. (B) Wild-type (■) or ARF-null (░⃞) MEFs infected for 36 hr with the indicated viruses (abscissa) were labeled for 14 hr with BrdUrd and scored for protein expression and BrdUrd incorporation as in Fig. 4. (C) NIH 3T3 cells were cotransfected with the ARF promoter-reporter plasmid together with vectors encoding E2F-1 or both E2F-1 and DMP1. Input plasmid DNAs (μg) are noted on the abscissa and activation was normalized to coexpressed SEAP (ordinate). (D) Cells infected as in B were deprived of serum for 24 hr and then scored for viability by trypan blue exclusion. Viability was confirmed using fluorescence-activated cell sorter/terminal deoxynucleotidyltransferase-dUTP end labeling assay and by scoring representative aliquots for subdiploid DNA content.
E2F-1, E1A, c-Myc, and v-Abl induce p19ARF expression, but each also triggers the expression of genes that promote both G1 phase progression and apoptosis (16–19). Myc and E2F-1 acutely increased the S phase fraction of MEFs after viral infection, and unlike DMP1, neither exhibited differential effects in ARF-positive versus ARF-null MEFs maintained in the presence of serum (Fig. 3B). The mouse ARF promoter contains at least two potential E2F-1-binding sites in the −208 to −127 segment that includes the DMP1-binding site (Fig. 1). We confirmed results obtained with human ARF demonstrating that E2F-1 could stimulate ARF promoter-dependent gene expression (18) and also could act in conjunction with DMP1 (Fig. 3C). The human ARF promoter also contains a high-affinity DMP1-binding site (GACGGATGT) at nucleotides −397 to −389, relative to the transcriptional start site (data not shown). As for Myc, the growth-promoting effects of E2F-1 were sufficient to override ARF-induced arrest (Fig. 3B).
On the other hand, MEFs overexpressing c-Myc, E2F-1, or E1A are exquisitely sensitive to apoptosis. The ability of these proteins to induce cell death is enhanced in MEFs deprived of serum (36) but is significantly attenuated in cells lacking ARF or p53 function (16, 17). Importantly, ARF overexpression per se does not trigger apoptosis in MEFs (14), so the proapoptotic functions of Myc or E2F-1, although countered by ARF loss, are likely mediated through other target genes. In wild-type MEFs deprived of serum, ectopic Myc expression induced cell death, but DMP1, like ARF, led to growth arrest and did not trigger apoptosis (Fig. 3D), as confirmed by fluorescence-activated cell sorter/terminal deoxynucleotidyltransferase-dUTP end labeling assays (data not shown).
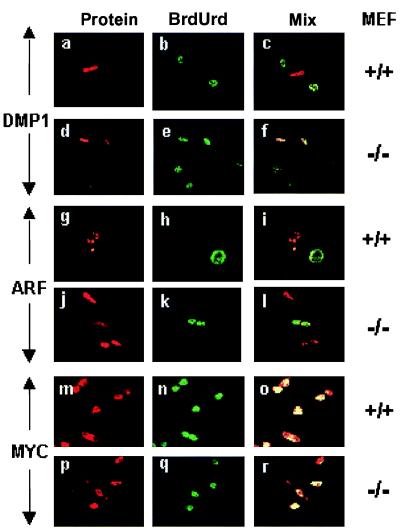
Fig. 4 illustrates representative data obtained with both wild-type and ARF-null MEFs infected with DMP1 virus (Fig. 4 a–f), ARF virus (Fig. 4 g–l), or c-Myc virus (Fig. 4 m–r). Both ectopically expressed DMP1 and ARF induced cell cycle arrest. Cells expressing either of these proteins (red fluorescence) did not incorporate BrdUrd (green fluorescence), whereas uninfected cells in the same cultures proceeded into S phase. Although both wild-type (Fig. 4 g–i) and ARF-null cells (Fig. 4 j–l) were arrested by ectopically expressed ARF protein, DMP1 was effective only in ARF-positive cells (Fig. 4 a–c). In contrast, cells infected with c-Myc virus continued to proliferate when maintained in serum-containing medium (Fig. 4 m–r).
Figure 4.
DMP1-induced arrest depends on ARF. Wild-type or ARF-null MEFs (Right) were infected for 36 hr with the different expression vectors (Left) and scored for vector-induced protein expression (red fluorescence, Left), BrdUrd incorporation (green fluorescence, Center), and mixed fluorescence (yellow, Right). Wild-type DMP1-overexpressing cells failed to incorporate BrdUrd (a–c), whereas ARF-null cells entered S phase (d–f). ARF arrests both MEF cell types (g–l) and Myc arrests neither (m–r).
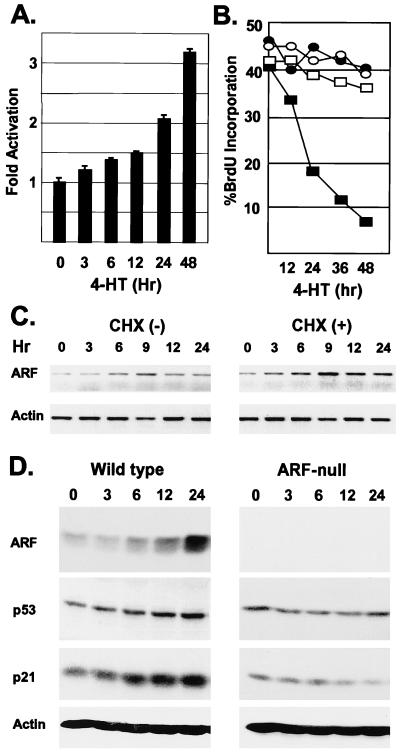
To determine the kinetics of ARF induction in response to DMP1, we created a DMP1-ER fusion protein that is conditionally regulated in response to 4-HT. In NIH 3T3 fibroblasts, the DMP1-ER construct activated a cotransfected ARF-luciferase promoter-reporter construct in response to 4-HT treatment (Fig. 5A). When wild-type primary MEFs infected with a DMP1-ER retrovirus were treated with 4-HT, they underwent growth arrest, whereas ARF-null MEFs did not respond (Fig. 5B). 4-HT treatment of wild-type cells induced expression of ARF mRNA (Fig. 5C) and protein (Fig. 5D). The increase in ARF mRNA was maximal by 9 hr after 4-HT addition and was potentiated when cells were also treated with cycloheximide (Fig. 5C), indicating that DMP1-mediated induction does not require new protein synthesis. 4-HT treatment led to increases in p53 and the p53-responsive CDK inhibitor, p21Cip1, whereas ARF-null MEFs did not exhibit increases in either protein (Fig. 5D), consistent with results above indicating that DMP1-induced arrest depends on ARF function. In turn, ARF-induced arrest strictly depends on functional p53 (20), and as expected, the proliferation of MEFs derived from p53-null mice was not affected by DMP1 (negative data not shown).
Figure 5.
Conditional ARF induction and growth arrest of wild-type MEFs by DMP1-ER. (A) NIH 3T3 cells treated with 4-HT for the indicated times (hr, abscissa), were scored for activation (normalized to SEAP) of a cotransfected ARF-promoter-reporter plasmid. (B) Wild-type (■, ●) or ARF-null (○, □) MEFs expressing DMP1-ER were left untreated (○, ●) or were treated with 4-HT (□, ■) for the indicated times (abscissa). Cells were pulsed with BrdUrd for 3 hr before analysis. (C) ARF and actin mRNA were quantitated by reverse transcription–PCR in lysates of wild-type cells treated with 4-HT as in B or with 4-HT plus the protein synthesis inhibitor cycloheximide (CHX). (D) ARF, p53, p21Cip1, and actin protein levels were determined by immunoblotting in lysates of wild-type (Left) and ARF-null (Right) MEFs treated with 4-HT as in B.
DISCUSSION
The DMP1 transcription factor binds to a single consensus site in the mouse ARF promoter to activate gene expression. Mutation of this binding site abolished DMP1-stimulated expression of a reporter gene driven by a DNA fragment containing residues −225 to +56 of the ARF promoter. Conversely, a DMP1 point mutant that no longer binds to DNA was transcriptionally inert. Ets1 and Ets2 transcription factors can also bind to short oligonucleotides containing the DMP1 consensus binding site, but five Ets family members were unable to activate reporter gene expression driven by the larger 281-bp ARF promoter fragment, suggesting that DMP1 may be the preferred regulator in this context. We have seen similar effects with the promoter of the aminopeptidase-N/CD13 gene on which DMP1-DNA complexes were significantly more stable than those containing Ets factors (32). In agreement with these findings, ectopic expression of DMP1 in wild-type MEF strains induced expression of the p19ARF protein and caused cell cycle arrest, but Ets1 overexpression was without effect.
Mutants of DMP1 defective in DNA binding or in transactivation do not cause cell cycle arrest, underscoring the requirement for target gene expression (27). The fact that ARF-null MEFs did not stop dividing in response to DMP1 now provides direct evidence that ARF function is required for DMP1’s antiproliferative effects on the cell cycle, at least in primary diploid fibroblasts. ARF-induced arrest depends on p53 (20), and conditional activation of a DMP1-ER fusion protein induced expression of p53 and of the p53-regulated p21Cip1 protein in wild-type MEFs, but not in their ARF-null counterparts. As expected, the proliferation of p53-null MEFs was also unaffected by DMP1, and p21Cip1 was not induced (data not shown). Therefore, DMP1 up-regulates ARF gene expression in primary MEFs, leading in turn to p53-dependent growth arrest.
These data do not formally preclude the possibility that DMP1 can also activate other genes important in cell cycle control. For example, much higher levels of ectopic DMP1 expression can inhibit cell cycle entry in NIH 3T3 cells that have sustained ARF deletions, implying that DMP1 may coregulate other relevant targets. Myc can activate p53 through ARF-dependent and ARF-independent pathways, although much higher Myc levels are required to activate p53 when ARF is absent (16). As shown here, however, DMP1 does not induce p53 in ARF-null MEFs. In a survey for other DMP1-regulated genes, we observed that DMP1 can indirectly up-regulate reporter gene expression from the p27Kip1 promoter in NIH 3T3 cells (32), even though this promoter fragment lacks a consensus DMP1 binding site. Unlike ARF-null cells, the proliferation of p27Kip1-null MEFs is still inhibited by DMP1 (data not shown). DMP1 did not induce p21Cip1 in ARF-null MEFs (Fig. 5D), and did not significantly induce reporter gene expression driven by the p21Cip1 promoter (data not shown).
Cell cycle arrest induced by ectopic expression of DMP1 is antagonized by D-type cyclin overexpression (27). In agreement, we have also observed that coexpression of cyclin D1 with DMP1-ER in primary MEFs can counter cell cycle arrest induced by 4-HT (data not shown). Effects of D-type cyclins on DMP1-mediated gene expression could occur as a result of direct physical interactions between DMP1 and D-type cyclins or, alternatively, through CDK4-dependent phosphorylation of the retinoblastoma protein and up-regulation of E2Fs, which can drive cells into S phase.
The effects of DMP1 on the ARF–p53 pathway differ in several key respects from the consequences of overexpression of Myc, E1A, E2F-1, all of which also activate p19ARF synthesis in MEFs (16–18). First, Myc has not been demonstrated to bind to the ARF promoter, and its inductive effects on p19ARF protein synthesis may well be indirect. Second, ectopic Myc and E2F-1 expression cause conflicting biologic responses in the sense that both stimulate S phase entry and yet trigger programmed cell death. The apoptotic response can be masked by survival factors that are normally present in cell culture medium, so that cell death becomes more pronounced when MEFs overexpressing Myc are deprived of serum (36). Compared with normal cells, those that have lost ARF or p53 function become relatively resistant to apoptosis induced by Myc, and such variants soon become established as continuously proliferating cell lines (16). In the latter populations, the growth-promoting effects of Myc are unchecked, and the proliferative rate of the cells is accelerated. Therefore, Myc and E2F overexpression is normally countered by ARF-dependent signals that antagonize rapid cell proliferation and help to promote apoptosis in a p53-dependent manner (reviewed in ref. 15). Because enforced ARF expression arrests wild-type MEFs but does not kill them (14), other functions of Myc in addition to activation of the ARF-p53 pathway are required for apoptosis. DMP1 lacks these collateral functions, because like ARF itself, DMP1 induces cell cycle arrest but does not provoke cell death, at least in this setting.
Because the ARF and DNA damage pathways that impinge on p53 are distinct, activation of ARF by low levels of Myc or E1A can sensitize cells to the p53-dependent effects of genotoxic drugs or irradiation (17). However, the growth-promoting properties of Myc and E1A also contribute to rapid selection of drug-resistant variants that lose ARF–p53 checkpoint control (16). Because DMP1 exhibits no overt growth-promoting functions, it may be more useful as a specific sensitizer of p53-dependent killing in response to common chemotherapeutic regimens, without as great a risk of selection for p53-negative variants.
Acknowledgments
We thank Jacques Ghysdael, Linda Shapiro, Scott Hiebert, Jacqueline Lees, and Charles Sawyers for human Ets family, E2F-1, and c-Myc cDNAs; Takehiko Kamijo and Frederique Zindy for MEF cell strains; Richard A. Ashmun for flow cytometric analyses; and Esther Van de Kamp and Rose Matthew for excellent technical assistance. C.J.S. is an Investigator of the Howard Hughes Medical Institute. This work was supported in part by National Institutes of Health Grants CA-56819 and CA-71907 (to M.F.R.), by Cancer Center Core Grant CA-21765, and by the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital.
ABBREVIATIONS
- MEFs
mouse embryonic fibroblasts
- 4-HT
4-hydroxytamoxifen
- SEAP
secreted alkaline phosphatase
- ER
estrogen receptor
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF120108).
References
- 1.Hollstein M, Rice K, Greenblatt M S, Soussi T, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montesano R, Harris C C. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 2.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 3.Prives C. Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 4.Barak Y, Juven T, Haffner R, Oren M. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, Bayle J H, Olson D, Levine A J. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 6.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 7.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Nature (London) 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 8.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 9.Honda R, Tanaka H, Yasuda H. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 10.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 11.Roth J, Dobbelstein M, Freedman D, Shenk T, Levine A J. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruas M, Peters G. BBA Rev Cancer. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 13.Serrano M, Hannon G J, Beach D. Nature (London) 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 14.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 15.Sherr C J. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 16.Zindy F, Eischen C M, Randle D, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Stanchina E, McCurrach M E, Zindy F, Shieh S-Y, Ferbeyre G, Samuelson A V, Prives C, Roussel M F, Sherr C J, Lowe S W. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bates S, Phillips A C, Clarke P, Stott F, Peters G, Ludwig R L, Vousden K H. Nature (London) 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 19.Radfar A, Unnikrishnan I, Lee H-W, DePinho R A, Rosenberg N. Proc Natl Acad Sci USA. 1998;95:13194–13199. doi: 10.1073/pnas.95.22.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 21.Stott F, Bates S A, James M, McConnell B B, Starborg M, Brookes S, Palmero I, Hara E, Vousden K H, Peters G. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pomerantz J, Schreiber-Agus N, Liégeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, et al. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Xiong Y, Yarbrough W G. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 24.Kamijo T, Weber J D, Zambetti G, Zindy F, Roussel M F, Sherr C J. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliner J D, Kinzler K W, Meltzer P S, George D L, Vogelstein B. Nature (London) 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 26.Hirai H, Sherr C J. Mol Cell Biol. 1996;16:6457–6467. doi: 10.1128/mcb.16.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue K, Sherr C J. Mol Cell Biol. 1998;18:1590–1600. doi: 10.1128/mcb.18.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bodner, S. M., Naeve, C. W., Rakestraw, K. M., Jones, B. G., Valentine, V. A., Valentine, M. B., Luthardt, F. W., Willman, C. L., Raimondi, S. C., Downing, J. et al. (1999) Gene, in press. [DOI] [PubMed]
- 29.Zwijsen R M L, Wientijens E, Klompmaker R, van der Sman J, Bernards R, Michalides R J A M. Cell. 1997;88:405–415. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]
- 30.Neuman E, Ladha M H, Lin N, Upton T M, Miller S J, Direnzo J, Pestell R G, Hinds P W, Dowdy S F, Brown M, Ewen M E. Mol Cell Biol. 1997;17:5338–5347. doi: 10.1128/mcb.17.9.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganter B, Fu S L, Lipsick J S. EMBO J. 1998;17:255–268. doi: 10.1093/emboj/17.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue K, Sherr C J, Shapiro L H. J Biol Chem. 1998;273:29188–29194. doi: 10.1074/jbc.273.44.29188. [DOI] [PubMed] [Google Scholar]
- 33.Zindy F, Quelle D E, Roussel M F, Sherr C J. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 34.Davis N, Roussel M F. Gene. 1996;171:265–269. doi: 10.1016/0378-1119(96)00013-3. [DOI] [PubMed] [Google Scholar]
- 35.Bailly R A, Bosselut R, Zucman J, Cormier F, Delattre O, Roussel M F, Thomas G, Ghysdael J. Mol Cell Biol. 1994;14:3230–3241. doi: 10.1128/mcb.14.5.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Water C M, Penn L Z, Hancock D C. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]