Abstract
Estrogen receptor (ER) modulators produce distinct tissue-specific biological effects, but within the confines of the established models of ER action it is difficult to understand why. Previous studies have suggested that there might be a relationship between ER structure and activity. Different ER modulators may induce conformational changes in the receptor that result in a specific biological activity. To investigate the possibility of modulator-specific conformational changes, we have applied affinity selection of peptides to identify binding surfaces that are exposed on the apo-ERs α and β and on each receptor complexed with estradiol or 4-OH tamoxifen. These peptides are sensitive probes of receptor conformation. We show here that ER ligands, known to produce distinct biological effects, induce distinct conformational changes in the receptors, providing a strong correlation between ER conformation and biological activity. Furthermore, the ability of some of the peptides to discriminate between different ER α and ER β ligand complexes suggests that the biological effects of ER agonists and antagonists acting through these receptors are likely to be different.
The estrogen receptor (ER) is a member of the steroid family of nuclear receptors. Like other nuclear receptors, the ER is a ligand-dependent transcriptional activator (1). In the absence of hormone, the ER resides in the nucleus of target cells where it is associated with an inhibitory heat-shock protein complex (2). On binding ligand, the receptor is activated. This process permits the formation of stable receptor dimers and subsequent interaction with specific DNA response elements located within the regulatory region of target genes (3). The DNA-bound receptor can then either positively or negatively regulate target gene transcription. Although the precise mechanism by which the ER modulates RNA polymerase activity remains to be determined, it has been shown recently that agonist-bound ER can recruit transcriptional adaptors, proteins that permit the receptor to transmit its regulatory information to the cellular transcriptional apparatus (4–6). Conversely, when occupied by antagonists, the DNA-bound receptor actively recruits corepressors, proteins that permit the cell to distinguish between agonists and antagonists (5–7). Building on this complexity was the recent discovery of a second ER, ER β, whose mechanism of action appears to be similar to, yet distinct from, ER α (8–10).
Drugs that target the ER can exhibit a variety of effects in different target tissues. For example, tamoxifen is an ER antagonist in breast tissue (11) but an ER agonist in bone (12) and uterine (13) tissue. Raloxifene is also an ER antagonist in breast tissue; however, it exerts agonist activity in bone but not uterine tissue (14). Indeed, one of the greatest challenges in understanding the pharmacology of the ER is determining how different ER ligands produce such diverse biological effects. We have explored the possibility that various ER ligands induce distinct conformational changes in the ER. These distinct conformations may, in turn, alter the interactions of the receptor with cell- and tissue-specific coactivating or corepressing proteins or even estrogen response elements (EREs), thus leading to diverse biological effects. Using limited proteolysis, we and others have shown that the ER agonist estradiol and the ER antagonist ICI 182,780 induced distinct ER conformations (15, 16). However, the picture is much more complicated than this. There is a variety of ER ligands, selective ER modulators, which are neither pure agonists nor antagonists. These ligands, which include tamoxifen and raloxifene, produce distinct tissue-specific biological effects, yet conformational differences cannot be discerned in the protease digestion assay (15, 16). It is likely that these compounds are also eliciting distinct conformational changes that affect ER activity, but the changes are too subtle to be detected by the protease digestion assay (17, 18).
In an effort to explore the relationship between ER structure and biological activity, we have used affinity-selected peptides to probe the conformational changes that occur within the ER on binding various ligands. Our results indicate that different peptide-binding surfaces on the ER are exposed in response to binding different ligands, and that these binding surfaces are distinct from those exposed on the apo-receptor. We infer from these results that different ER–ligand complexes may be able to contact different proteins within the cell, and that the overall biological response is determined by unique combinations of protein–protein interactions that occur in a given cell and promoter context.
MATERIALS AND METHODS
Materials.
ER α and β were purchased from Panvera (Madison, WI). Immulon 4 96-well plates were from Dynatech. Streptavidin, 17-β estradiol, 4-OH tamoxifen, nafoxidine, clomiphene, diethylstilbestrol, progesterone, 16-α OH estrone, and estriol were purchased from Sigma. Premarin is a product of Wyeth-Ayerst Laboratories (Marietta, PA). Raloxifene is a product of Eli Lilly. ICI 182,780 was purchased from Tocris Cookson (Ballwin, MO). Anti-M13 antisera was purchased from Amersham Pharmacia. Sequencing of single-strand M13 DNA was conducted by Sequetech (Mountain View, CA). Peptide synthesis was conducted by AnaSpec (San Jose, CA). Oligonucleotides corresponding to the vitellogenin ERE, biotin- GATCTAGGTCACAGTGACCTGCG (forward) and biotin-GATCCGCAGGTCACTGTGACCTA (reverse), were synthesized by Genosys (The Woodlands, TX).
Phage Affinity Selection.
Affinity selection of phage for the various conformations of the ER was conducted essentially as described (19). Immulon 4 96-well plates were coated with streptavidin in 0.1 M sodium bicarbonate. The plates were then incubated for 1 h with 2 pmol biotinylated vitellogenin ERE per well (20), followed by incubation for 1 h with 3 pmol (monomer) ER α or ER β per well. Affinity selections were conducted with the ER in TBST (10 mM Tris⋅HCl, pH 8.0/150 mM NaCl/0.05% Tween 20) or in TBST containing 1 μM 17-β estradiol or 4-OH tamoxifen.
Phage ELISA.
ER α or β was immobilized on the vitellogenin ERE as described for phage affinity selection. The ER was then incubated with 100 μl TBST or TBST containing the appropriate modulator. Phage (40 μl) from a 5-h culture grown in DH5αF′ cells was added directly to the wells and incubated 30 min at room temperature. Unbound phage were then removed by five washes with TBST. Bound phage were detected by using an anti-M13 antibody coupled to horseradish peroxidase. Assays were developed with 2,2-azinobis(3-ethylbenzothiazoline)-6 sulfonic acid and hydrogen peroxide for 10 min and then stopped by the addition of 1% SDS. Absorbance was measured at 405 nm in a Molecular Devices microplate reader.
Mapping Phage Binding Sites on ER α.
For mapping studies, the ER α ligand-binding domain (residues 282–595) fused to glutathione S-transferase (GST) (a gift from Peter Kushner) and the ER α N terminus (residues 1–184) fused to GST were used. The full length ER α or domains were immobilized directly on the surface of the Immulon 4 plate. Assays were conducted as described for phage ELISA.
Time-Resolved Fluorescence Assays.
Time-resolved fluorescence (TRF) assays were performed at room temperature as follows: costar high-binding 384-well plates were coated with streptavidin in 0.1 M sodium bicarbonate and blocked with BSA. Biotinylated ERE (2 pmol) was added to each well. After a 1-h incubation, biotin was added to block any remaining binding sites. The plates were washed, and 2 pmol ER α was added to each well. Following a 1-h incubation, the plates were washed and the ER modulators were added at a range of concentrations. Following a 30-min incubation with the modulators, 2 pmol of a europium-labeled streptavidin (Wallac, Gaithersburg, MD) -biotinylated peptide conjugate (prepared as described below) was added and incubated for 1 h. The plates were then washed and the europium enhancement solution was added. Fluorescent readings were obtained with a POLARstar fluorimeter (BMG Lab Technologies) by using a <400-nm excitation filter and a 620-nm emission filter. The europium-labeled streptavidin-biotinylated peptide conjugate was prepared by adding 8 pmol biotinylated peptide to 2 pmol labeled streptavidin. After incubation on ice for 30 min, the remaining biotin-binding sites were blocked with biotin before addition to the ER-coated plate.
RESULTS AND DISCUSSION
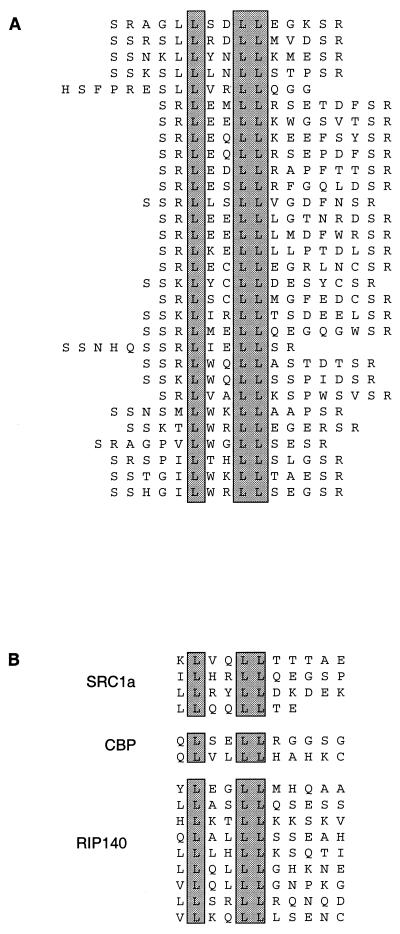
Affinity selection of phage-displayed peptide libraries (19) was conducted on both ER α and β under conditions that were predicted to place the ER in different conformations — apo-ER, estradiol-bound ER, and 4-OH tamoxifen-bound ER. Unique sets of high-affinity peptides were identified under each condition. Most notably, affinity selection of peptides in the presence of estradiol revealed a number of sequences containing an LXXLL motif (Fig. 1A). This motif, which is found in nuclear receptor coactivators (Fig. 1B), has been shown to be necessary and sufficient for their association with nuclear receptors (21). Studies have shown that the association of the LXXLL motif with the ER is accomplished via a helical region in the ligand-binding domain of the receptor that is exposed on binding estradiol. Structural studies using x-ray crystallography have shown that this region is not properly positioned in the presence of raloxifene (17) or 4-OH tamoxifen (18), thus preventing the interaction of the coactivator LXXLL motif. The identification of these sequences in the presence of estradiol indicates that the ER is undergoing conformational changes in response to ligand in vitro consistent with the changes that are predicted to occur in vivo.
Figure 1.
(A) Sequences of LXXLL motif containing peptides that were affinity selected on ER α in the presence of estradiol. (B) Sequences of LXXLL motifs found in the nuclear receptor coactivating proteins human SRC1a (steroid receptor coactivator 1a), mouse cAMP-responsive element binding protein (CREB)-binding protein (CBP), and human RIP140.
All of the affinity-selected phage were evaluated by phage ELISA for binding to apo-ER α and β and to ER α and β in the presence of estradiol or 4-OH tamoxifen, as described in Materials and Methods and illustrated in Fig. 2. Many phage showed distinct preferential binding. Some sequences bound more strongly to the apo-receptor, while others exhibited preferential binding to the estradiol-activated or the 4-OH tamoxifen-activated receptor. No signal was observed when the assays were carried out in the absence of ER, indicating that the peptides were binding to the ER ligand complex. Based on this analysis, 11 phage expressing different peptide sequences and showing distinct binding preferences were chosen for further use as conformational probes (Fig. 3). Five of the probes have affinity for both ER α and ER β and were designated α/β I–V. Three probes were specific for ER α, designated α I–III, and three were specific for ER β, designated β I–III. The identification of distinct classes of peptides, some of which recognized both ER α and ER β and others that were receptor specific, is consistent with the primary structures of the two receptors being similar yet distinct.
Figure 2.
Phage ELISA. A biotinylated vitellogenin ERE was immobilized on 96-well plates precoated with streptavidin. The ER was then immobilized on the ERE and incubated for 5 min in the presence of modulator before the addition of phage. Assays were conducted as described in Materials and Methods. HRP, horseradish peroxidase.
Figure 3.
Analysis of the binding specificity of the conformational probes was conducted by phage ELISA as described in Materials and Methods. Estradiol and 4-OH tamoxifen concentrations were 1 μM. The probes α/β I-α/β V are shown only for ER α. The binding patterns of these probes on ER β were similar. Sequences of the probes are as follows: α/β I, SSNHQSSRLIELLSR; α/β II, SAPRATISHYLMGG; α/β III, SSWDMHQFFWEGVSR; α/β IV, SRLPPSVFSMCGSEVCLSR; α/β V, SSPGSREWFKDMLSR; α I, SSEYCFYWDSAHCSR; α II, SSLTSRDFGSWYASR; α III, SRTWESPLGTWEWSR; β I, SREWEDGFGGRWLSR; β II, SSLDLSQFPMTASFLRESR; β III, SSEACVGRWMLCEQLGVSR.
The binding sites of the eight probes, α/β I–V and α I–III, were mapped on ER α by using isolated ER α ligand-binding domain, an amino terminal domain, and the full length ER. Assays were conducted by using the format shown in Fig. 2, except that the domains and the full length receptor were directly immobilized on the plastic surface of the well. All of the probes, except α I, bound to the ligand-binding domain. The α I probe, which binds only to the full length protein, may be binding to a site that is created by the tertiary structure formed by the interaction between receptor domains (data not shown). Whereas the binding of the probes requires the presence of the ER ligand-binding domain, we cannot at this time formally exclude the possibility that the binding surface for some probes is created by the combination of ligand and receptor. However, crystal structures of the ER ligand-binding domain complexed with ligand indicate that helix 12 is positioned over the ligand-binding site such that the probes may be sterically hindered from binding directly to the ligand (17, 18).
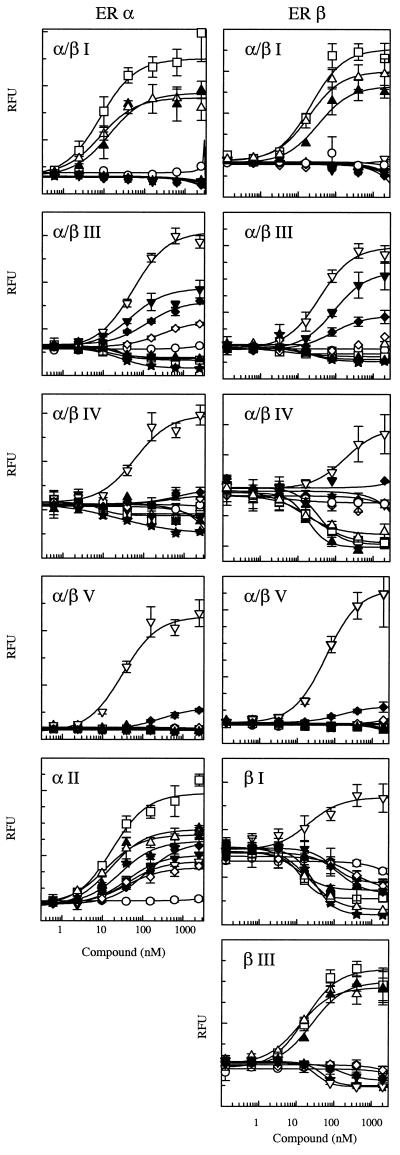
Next, we evaluated the binding of each of the probes to ER α and ER β in the presence of a variety of ER ligands that have distinct biological activities (22–31). The goal was to determine whether each of the ligands would induce a conformational change in the ER that would alter the binding pattern of the probes, thus producing a “fingerprint” for each compound. The ligands used for this study include the ER agonists estradiol, estriol, and diethylstilbestrol (DES); the selective ER modulators 4-OH tamoxifen, nafoxidine, clomiphene, and raloxifene; the antagonist ICI 182,780; and the estradiol metabolite 16-α-OH estrone. Premarin, the mixture of conjugated estrogens used as estrogen replacement therapy, was also included, but it should be noted that many of the components of Premarin must be metabolically activated. Thus, their action may not be detected in this in vitro assay. Buffer only (apo-receptor) and progesterone were included as controls. As shown in Fig. 4, each of the ligands tested did indeed alter the binding pattern of the probes, producing a distinct fingerprint for each, whereas the pattern produced by progesterone was indistinguishable from that produced by buffer.
Figure 4.
Fingerprint analysis of ER modulators on (A) ER α and (B) ER β. Immobilized ER was incubated with estradiol (1 μM), estriol (1 μM), Premarin (10 μM), 4-OH tamoxifen (1 μM), nafoxidine (10 μM), clomiphene (10 μM), raloxifene (1 μM), ICI 182,780 (1 μM), 16α-OH estrone (10 μM), DES (1 μM), or progesterone (1 μM). Phage ELISAs were conducted as described in Materials and Methods.
The unique ligand-dependent binding patterns of the probes indicates that each ligand induces a receptor conformational change that differentially exposes peptide-binding surfaces. The binding patterns for estradiol and ICI 182,780 are distinct on both ER α and β, confirming the conformational change illustrated by the earlier protease digestion studies (15). The protease digestion assay, which relies on the location of cleavage sites for detection of conformational changes, could distinguish between conformational changes induced by estradiol and 4-OH tamoxifen or estradiol and ICI 182,780. However, it was unable to distinguish between changes induced by 4-OH tamoxifen and other ER modulators such as ICI 182,780 (15). The fingerprint assay, however, clearly indicates that unique peptide-binding surfaces are exposed on both ER α and β in the presence of 4-OH tamoxifen that are not exposed in the presence of ICI 182,780. Tamoxifen, nafoxidine, and clomiphene contain the same triphenylethylene core structure. These three compounds, although similar in structure, produce distinct biological effects. Therefore, it might be predicted that these compounds would induce similar, yet distinct, conformational changes in the receptors. The fingerprint assay shows that the probes α/β III, IV and V, which have high affinity for the ER in the presence of 4-OH tamoxifen, have lower affinity for the ER complexed with nafoxidine and clomiphene, indicating that the exposure of these peptide-binding surfaces differs in the presence of these compounds. The α III probe more clearly differentiates these three compounds. The fingerprint assay also differentiates 4-OH tamoxifen and raloxifene. The probes α/β III, IV, and V have reduced affinity for both ER α and β in the presence of raloxifene compared with 4-OH tamoxifen. The probes α/β II, β I, and β III further distinguish ER β conformational changes induced by these two compounds. The fingerprint pattern produced by Premarin is distinct compared with other agonists; however, Premarin’s activities are caused by a mixture of components. It would be interesting to assess the binding patterns of the probes in the presence of each of the purified activated components of Premarin.
To confirm that the binding of the probes to the ER depended on the peptide expressed on the surface of the phage, biotinylated peptides corresponding to the phage sequences were synthesized with biotin attached to a C-terminal lysine. The peptides were coupled to europium-labeled streptavidin and binding studies were conducted by using TRF spectroscopy, as described in Materials and Methods. The concentrations of the modulators were varied from picomolar to micromolar, and the binding of these probes to the ER was measured. The results, shown in Fig. 5, indicate that the peptides are indeed conferring the binding specificity. Comparison of the fluorescence values obtained from the TRF binding assays and the signals obtained in the phage ELISA fingerprint indicate that the two methods produce similar patterns. However, the binding assay also provides an indication of the potency of each compound to induce the conformational change required for peptide binding (Table 1). Taken together, these results indicate that conversion of the fingerprint assay from phage to peptides will provide an even more sensitive assay for detecting conformational change.
Figure 5.
Comparison of the binding of the peptide probes to ER α or ER β in the presence of modulators using TRF. Assays were conducted as described in Materials and Methods. (○), buffer, aporeceptor; (□) 17β estradiol;(▵) estriol; (▴) DES; (▿) 4-OH tamoxifen; (▾) raloxifene; (⋄)nafoxidine; (♦) clomiphene; (★) ICI 182,780. RFU, relative fluorescence units.
Table 1.
Binding of the peptide probe to ER α and ER β in the presence of modulators
| Modulator | EC50 for ER peptide probe
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ER α
|
ER β
|
||||||||||
| α/β I | α/β III | α/β IV | α/β V | α II | α/β I | α/β III | α/β IV | α/β V | β I | β III | |
| Buffer | |||||||||||
| 17β-Estradiol | 8 | 18 | 8 | 17 | 22 | 6 | 27 | 13 | 17 | ||
| Estriol | 8 | 19 | 45 | 12 | 20 | 16 | 12 | 21 | 12 | ||
| 4-OH tamoxifen | 55 | 60 | 31 | 42 | 37 | 180 | 50 | 21 | 34 | ||
| Nafoxidine | 290 | 370 | 39 | 230 | 320 | ||||||
| Clomiphene | 140 | 710 | 280 | 120 | 82 | 150 | 140 | 120 | |||
| Raloxifene | 49 | 42 | 90 | 90 | 160 | ||||||
| ICI 182, 780 | 26 | 25 | 29 | 18 | 35 | 29 | 48 | ||||
| Diethylstilbesterol | 13 | 30 | 16 | 34 | 15 | 18 | 11 | 25 | |||
EC50 is defined as the concentration, in nanomolar, of a given modulator required to achieve a 50% change in the binding of the probe to the receptor. The change in conformation may result in an increase or decrease in the affinity of the probe for the receptor.
One of the most notable observations from the TRF binding assays is that the binding of the β I probe to ER β is enhanced in the presence of the selective ER modulator 4-OH tamoxifen and reduced in the presence of other SERMs such as raloxifene, nafoxidine, and clomiphene. The reduction in binding observed with these compounds is similar to the reduction observed with agonists such as estradiol, estriol, and DES.
We have identified peptides that serve as conformational probes of the ER α and β. Many probes bind to both receptors, while other probes bind to either the α or β receptor. Consistent with the two receptors having regions of high homology and other more divergent regions, these results indicate that the receptors have some binding surfaces in common, while others are unique. The implications of this are that both receptors may contact some of the same regulatory proteins in the cell, yet there may be additional proteins that specifically regulate either ER α or β action.
We have used our peptidic probes to show that both receptors undergo distinct conformational changes as a result of binding different ligands. The probes not only reveal receptor conformational changes by their relative changes in affinity, but they also identify unique binding surfaces on the two receptors. These binding surfaces may, in fact, be the surfaces that interact with various coregulatory proteins in response to different ligands. For example, many peptides selected with the estradiol-activated receptor contained sequences found in nuclear receptor coactivators, as illustrated by the peptides containing the LXXLL motif (Fig. 1). These peptide probes are probably mimicking the interaction between the receptor and coactivating proteins. Potentially, these probes can be used to identify heretofore unknown receptor–protein interactions.
Additional applications of the probes lie in the area of detection of ER modulators. One or more probes can be used to set up a high-throughput screen to identify modulators of ER activity. We anticipate that compounds that bind to the ER will alter receptor conformation and hence alter the binding patterns of the probes. The sites targeted by the screen may not be bona fide protein–protein interaction surfaces, but may represent sites exposed in the presence of a specific ligand and thus serve as markers for specific conformations. The fingerprinting technique may also be applied to quickly classify hits from a screen into different categories such as agonist (resembling the estrogen pattern), antagonist (resembling the ICI 182,780 pattern), mixed (resembling the tamoxifen pattern), or novel effectors, before assessing them in a cell-based assay. Fingerprinting may also be used to determine structure activity relationships and to rapidly assess compounds after chemical modification during lead optimization.
This is, to our knowledge, the first technique described that can distinguish between ER conformations induced by ligands both between and within ligand classes. The data gathered with this assay provide strong evidence that the biological activity of the ER can be linked to the conformation induced on binding ligand. A strength of this fingerprinting technique is that it is broadly applicable to any protein or receptor that undergoes structural changes on binding of a ligand or substrate. We are currently applying this technique to additional receptors and signaling proteins to aid in assessing conformational changes in response to chemical modulators of activity.
Acknowledgments
Some of this work was supported by a National Institutes of Health grant to D.P.M. (DK48807). We thank Bill Checovich and colleagues at Panvera Corp. for advice and support during this project, Brian Kay (University of Wisconsin, Madison, WI) for critical review of the manuscript, and Brett Antonio for technical assistance.
ABBREVIATIONS
- ER
estrogen receptor
- ERE
estrogen response element
- TRF
time-resolved fluorescence
- DES
diethylstilbestrol
References
- 1.Evans R M, Hollenberg S M. Cell. 1988;52:1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- 2.Smith D F, Toft D O. Mol Endocrinol. 1993;7:4–11. doi: 10.1210/mend.7.1.8446107. [DOI] [PubMed] [Google Scholar]
- 3.McDonnell D P, Nawaz Z, O’Malley B W. Mol Cell Biol. 1991;11:4350–4355. doi: 10.1128/mcb.11.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onate S A, Tsai S, Tsai M-J, O’Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 5.Norris J D, Fan D, Stallcup M R, McDonnell D P. J Biol Chem. 1998;273:6679–6688. doi: 10.1074/jbc.273.12.6679. [DOI] [PubMed] [Google Scholar]
- 6.Smith C L, Nawaz Z, O’Malley B W. Mol Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- 7.Lavinsky R M, Jepsen K, Heinzel T, Torchia J, Mullen T M, Schiff R, Del-Rio A L, Ricote M, Ngo S, Gemsch J, et al. Proc Natl Acad Sci USA. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greene G L, Gilna P, Waterfield M, Baker A, Hort Y, Shine J. Science. 1986;231:1150–1154. doi: 10.1126/science.3753802. [DOI] [PubMed] [Google Scholar]
- 9.Kuiper G G J M, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson J-Å. Proc Natl Acad Sci USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosselman S, Polman J, Dijkema R. FEBS Lett. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- 11.Jordan V C. Cancer. 1992;70:977–982. [PubMed] [Google Scholar]
- 12.Love R R, Mazess R B, Barden H S, Epstein S, Newcomb P A, Jordan V C, Carbone P P, DeMets D L. N Engl J Med. 1992;326:852–856. doi: 10.1056/NEJM199203263261302. [DOI] [PubMed] [Google Scholar]
- 13.Kedar R P, Bourne T H, Powles T J, Collins W P, Ashley S E, Cosgrove D O, Campbell S. Lancet. 1994;343:1318–1321. doi: 10.1016/s0140-6736(94)92466-x. [DOI] [PubMed] [Google Scholar]
- 14.Black L J, Sato M, Rowley E R, Magee D E, Bekele A, Williams D C, Cullinan G J, Bendele R, Kaufman F R, Bensch W R, et al. J Clin Invest. 1994;93:63–69. doi: 10.1172/JCI116985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonnell D P, Clemm D L, Hermann T, Goldman M E, Pike J W. Mol Endocrinol. 1995;9:659–669. doi: 10.1210/mend.9.6.8592512. [DOI] [PubMed] [Google Scholar]
- 16.Beekman J M, Allan G F, Tsai S Y, Tsai M-J, O’Malley B W. Mol Endocrinol. 1993;7:1266–1274. doi: 10.1210/mend.7.10.8264659. [DOI] [PubMed] [Google Scholar]
- 17.Brzozowski A M, Pike A C, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J-Å, Carlquist M. Nature (London) 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 18.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P, Agard D A, Greene G L. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 19.Sparks A B, Adey N B, Cwirla S, Kay B K. In: Phage Display of Peptides and Proteins, A Laboratory Manual. Kay B K, Winter J, McCafferty J, editors. San Diego: Academic; 1996. pp. 227–253. [Google Scholar]
- 20.Anderson I, Bartley C R, Lerch R A, Gray W G N, Friesen P D, Gorski J. Biochemistry. 1998;37:17287–17298. doi: 10.1021/bi981079b. [DOI] [PubMed] [Google Scholar]
- 21.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 22.Katzenellenbogen B S, Montano M M, Ekena K, Herman M E, McInerney E M. Breast Cancer Res Treat. 1997;44:23–38. doi: 10.1023/a:1005835428423. [DOI] [PubMed] [Google Scholar]
- 23.Macgregor J I, Jordan V C. Pharmacol Rev. 1998;50:151–196. [PubMed] [Google Scholar]
- 24.Zhu B T, Conney A H. Carcinogenesis. 1998;19:1–27. doi: 10.1093/carcin/19.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Subbiah M T R. Proc Soc Exp Biol Med. 1998;217:23–29. doi: 10.3181/00379727-217-44201. [DOI] [PubMed] [Google Scholar]
- 26.Sulistiyani, Adelman S J, Chandrasekaran A, Jayo J, St. Clair R W. Arterioscler Thromb Vasc Biol. 1995;15:837–846. doi: 10.1161/01.atv.15.7.837. [DOI] [PubMed] [Google Scholar]
- 27.Bhavnani B R, Cecutti A. J Clin Endocrinol Metab. 1994;78:197–204. doi: 10.1210/jcem.78.1.8288704. [DOI] [PubMed] [Google Scholar]
- 28.Bhavnani B. Proc Soc Exp Biol Med. 1998;217:6–16. doi: 10.3181/00379727-217-44199. [DOI] [PubMed] [Google Scholar]
- 29.Grese T A, Sluka J P, Bryant H U, Cullinan G J, Glasebrook A L, Jones C D, Matsumoto K, Palkowitz A D, Sato M, Termine J D, et al. Proc Natl Acad Sci USA. 1997;94:14105–14110. doi: 10.1073/pnas.94.25.14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grese T A, Pennington L D, Sluka J P, Adrian M D, Cole H W, Fuson T R, Magee D E, Phillips D L, Rowley E R, Shetler P K, et al. J Med Chem. 1998;41:1272–1273. doi: 10.1021/jm970688z. [DOI] [PubMed] [Google Scholar]
- 31.Howell, A (1997) Oncology (Basel)11, Suppl. 1, 59–64.