Abstract
Models of evolutionary processes postulate that new alleles appear in populations through random spontaneous mutation. Alleles that confer a competitive advantage in particular environments are selected and populations can be taken over by individuals expressing these advantageous mutations. We have studied the evolutionary process by using Escherichia coli cultures incubated for prolonged periods of time in stationary phase. The populations of surviving cells were shown to be highly dynamic, even after many months of incubation. Evolution proceeded along different paths even when the initial conditions were identical. As cultures aged, the takeovers by fitter mutants were incomplete, resulting in the coexistence of multiple mutant forms and increased microbial diversity. Thus, the study of bacterial populations in stationary phase provides a model system for understanding the evolution of diversity in natural populations.
In studies of microbial evolution, population takeovers have been observed in chemostats (1, 2) and by using serial transfer of batch cultures (3, 4). In both the chemostat and serial transfer systems, cells are continuously incubated in defined medium supplemented with limiting amounts of an essential nutrient to restrict growth rates and population sizes. These model systems create culture environments that are essentially constant, leading to selection of specific phenotypes (reviewed in ref. 5). Occasionally, a mutation arises that confers a selective advantage on a cell. This cell can outcompete its siblings and replace the population with its own progeny, in a process sometimes referred to as “periodic” selection (1, 5, 6). It is generally assumed that the result of these periodic selections is a complete takeover of the culture by cells of a single genotype.
We have chosen to study evolution of the bacterium Escherichia coli during extended stationary phase incubation by using a constant batch culture system in which no nutrients are added after the initial inoculation and no cells are removed from the system either by dilution (as in a chemostat) or by transfer to fresh medium (as in serial passage systems). This mode of incubation ensures that there is no loss of genetic diversity due to the arbitrary discarding of cells, removing the “bottlenecks” that can be incurred in other systems. By using this type of long-term batch culture system we had previously shown that cultures of a standard laboratory wild-type strain (ZK126) grown in rich medium saturate at ≈5 × 109 colony forming units (cfu) ml−1. Viability over the next few days decreases by about two orders of magnitude, stabilizing at ≈5 × 107 cfu ml−1. However, these surviving cells are different from their parents (7–10). Cells aged for 10 days in Luria–Bertani (LB) medium will take over a fresh overnight culture of the parental strain when the aged cells are introduced as a minority, in a phenomenon referred to as the growth advantage in stationary phase (GASP) phenotype (7–10). It was shown that the GASP phenotype is conferred by the acquisition of increased fitness mutations, as was the case in the chemostat and serial transfer experiments (1–5). Here again, it was assumed that a single mutant, which arose during periods of rapid growth, completely took over the population. Although all of the previous studies showed population takeovers by fitter mutants, none of them directly addressed two important questions in microbial evolution: (i) Given identical initial conditions does evolution always proceed along the same path? and (ii) Does the appearance of advantageous mutations always result in a single genotype dominating the population? By analyzing cultures incubated in stationary phase for much longer periods of time we have directly addressed these questions.
MATERIALS AND METHODS
Bacterial Strains, Culture Media, and Cell Growth Conditions.
Strains ZK1142 (NalR) and ZK1143 (StrR) are E. coli K-12 F− (nonmating) derivatives and have been described elsewhere (7). The nalidixic acid-resistance (NalR) and streptomycin-resistance (StrR) markers (encoded by gyrA and rpsL, respectively) are effectively neutral in the absence of drug selection, having no effect on competitive advantage under these growth conditions (7). In addition, we routinely assay for the spontaneous acquisition of both drug markers in either strain and have not observed a NalR strain become StrR, or vice versa. All bacterial cultures were incubated in 5.0 ml of LB broth (11) in 18 × 150 mm glass test tubes with constant aeration in a New Brunswick roller at 37°C.
Long-Term Growth of Bacterial Cultures.
Cultures of ZK1142 and ZK1143 have continued incubating for more than 1 year (data not shown) and titers remain at ≈106 cfu ml−1. Sterile distilled water is added to each culture monthly to adjust for the loss of volume because of evaporation.
Mixing of Cells from Cultures Aged 1–30 Days.
LB cultures (5 ml) of strains ZK1142 (NalR) and ZK1143 (StrR) were incubated at 37°C with aeration. Samples of each culture were taken after 1, 10, 20, and 30 days of constant incubation and stored as frozen LB/20% glycerol stocks at −80°C. For GASP competition experiments, fresh overnight LB cultures of each strain (from 1-, 10-, 20-, and 30-day-old cultures) were inoculated from frozen stocks. Mixes were done by transfer of 5 μl of cells from an overnight culture of the aged cells (as the minority) into a 5.0 ml overnight culture inoculated with cells from the younger culture. Viable counts were determined by serial dilution of cells removed periodically from the culture followed by plating on LB agar containing either nalidixic acid or streptomycin at 20 or 25 μg/ml, respectively (11).
Mixing Experiments Showing Population Dynamism.
Ten pairs of 5.0 ml cultures of ZK1142 (NalR) and ZK1143 (StrR) were incubated for 30 days at 37°C with aeration and then 2.5 ml each mixed to generate 10 new pairs of cultures. The time of mixing became Week 0, and mixed cultures were allowed to continue incubating. Viable counts of each subpopulation were determined biweekly as described above.
Mixing Experiments Showing Evolution Along Different Paths.
One culture of ZK1142 (NalR) and two cultures of ZK1143 (StrR-1 and StrR-2) were incubated for 30 days at 37°C with aeration. After 30 days, the NalR culture was mixed 1:1 with each of the cultures from the pair carrying the StrR drug marker. This resulted in two new cultures where each strain was mixed with two different, initially isogenic, aged cultures. Incubation of the mixed cultures was then continued and titers of NalR and StrR subpopulations were determined weekly as described above. This experiment was done twice in pairs, resulting in eight mixed cultures being tested.
Observation of Cells with Altered Colony Morphotypes.
Cells from long-term incubated batch cultures were plated periodically onto LB agar and incubated for 24–48 hr at 37°C. The number of colonies showing each morphotype was determined and individual colonies were picked and restreaked onto LB agar plates.
Preparation of Chromosomal DNA and Pulsed-Field Gel Electrophoresis.
Chromosomal DNA in agarose microbeads was prepared as described (12). Restriction enzymes were purchased from New England Biolabs and used according to the manufacturer’s instructions. Pulsed-field gels were run using 1% agarose gels in a Bio-Rad CHEF apparatus following the manufacturer’s instructions.
RESULTS
GASP Mutants Arise Constantly During Long-Term Incubation.
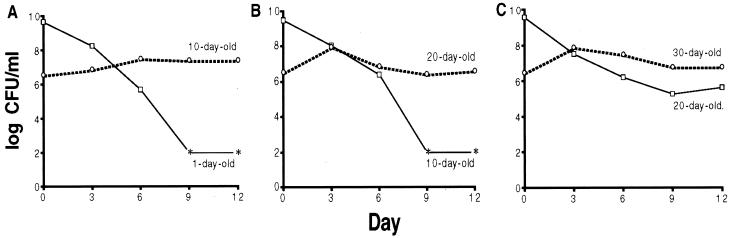
New GASP mutants are continuously selected as cultures incubate for extended periods of time. For example, not only do cells from 10-day-old cultures outcompete cells in fresh overnight cultures (Fig. 1A), but cells isolated from 20-day-old cultures can outcompete those from 10-day-old cultures (Fig. 1B), and mutants from 30-day-old cultures express a competitive advantage when mixed with cells from 20-day-old cultures (Fig. 1C). The observation that cells from progressively aged cultures have a competitive advantage over previous generations suggests that cells incubated under these conditions are under constant selection for the appearance of fitter mutants and implies that these long-term stationary phase cultures are highly dynamic, despite the fact that viable cell numbers remain roughly constant. Therefore, we designed experiments to directly observe the dynamism of these cultures using a series of mixing experiments with initially isogenic strains carrying either nalidixic acid (NalR) or streptomycin (StrR) resistance markers.
Figure 1.
Consecutive generations of GASP mutants arise in the same culture. Progressively aged cultures were mixed. (A) One-day-old in the majority (solid line) vs. 10-day-old in the minority (broken line). (B) Ten-day-old in the majority (solid line) vs. 20-day-old in the minority (broken line). (C) Twenty-day-old in the majority (solid line) vs. 30-day-old in the minority (broken line). Asterisks indicate that cfu ml−1 were below the limit of detection (<102 cfu ml−1).
Prolonged Dynamism of Stationary Phase Cultures.
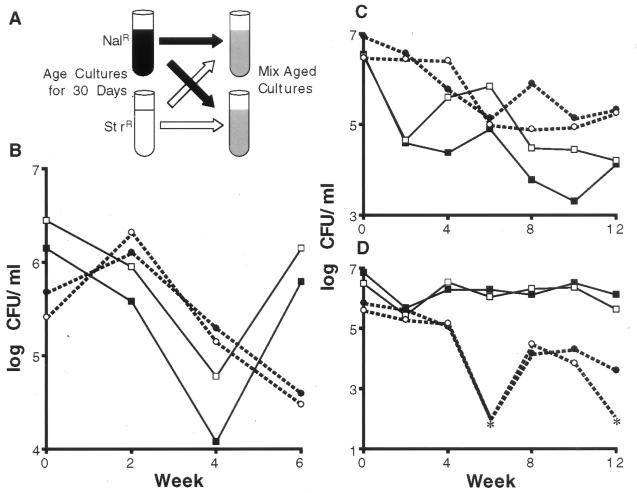
The results in Fig. 2 show that long-term stationary phase cultures are highly dynamic. Ten pairs of initially isogenic “founder” cultures (marked with either NalR or StrR) were aged for 30 days to allow for several cycles of GASP takeovers. These aged NalR and StrR cultures were then mixed in equal numbers, in duplicate (Fig. 2A). The titers of NalR and StrR cells were determined biweekly. In each of the 10 pairs of mixed cultures, the proportion of NalR and StrR cells varied over time, where the total viable counts remained roughly constant (≈106–107 cfu ml−1), directly demonstrating that these cultures are dynamic. An example of this population dynamism is shown in Fig. 2B. In this case, the number of StrR cells increases after 2 weeks, with a concomitant decrease in the number of NalR cells. This suggests that on mixing, GASP mutants in the StrR population were better competitors than the GASP mutants in the NalR culture. However, from weeks 4 to 6, the NalR subpopulation titer increased 100-fold with a concomitant decrease in the StrR cell numbers, indicating that the next GASP mutants able to take over the culture came from the NalR subpopulation. (In some cultures subpopulation titers changed more than four orders of magnitude in two weeks.)
Figure 2.
Dynamism in stationary phase cultures. (A) Five milliliter cultures of either nalidixic acid (NalR)- or streptomycin (StrR)-resistant bacteria were incubated for 30 days and then mixed 1:1 to generate two new cultures. (B–D) Three representative pairs of mixed NalR and StrR cells. Each culture of the pair is represented by either the solid or open symbols. NalR cells are represented by solid lines with squares, StrR cells are represented by broken lines with circles. Asterisks indicate that cfu ml−1 were below the limit of detection (<102 cfu ml−1).
The patterns of GASP takeovers in all 10 pairs of cultures during the first 2 weeks of incubation occurred in parallel. As shown in Fig. 2B, the points of inflection in the subpopulation titers occurred at the same time. This initial parallelism was expected because the long-term cultures were prepared in duplicate due to the transfer of one-half of each aged NalR or StrR culture into two newly mixed cultures (see Fig. 2A). The initial subpopulations in each pair of cultures should have been genetically identical. That is, the current best competitors comprising the majority of cells in the NalR and StrR 30-day-old founder cultures are present in both mixed cultures. Therefore, we expected that the patterns of cell growth and death, as reflected by changes in the relative titers of each marked subpopulation, would be similar between each pair of cultures shortly after mixing. Most pairs of cultures diverged between 2 and 6 weeks after initial mixing (Table 1). Fig. 2C shows a mixed pair in which parallel growth patterns are lost after 2 weeks of incubation.
Table 1.
Divergence of long-term cultures after 24 weeks of incubation
| Culture pair (A and B)* | Time of divergence† (weeks) | Drug marker(s) remaining‡
|
|
|---|---|---|---|
| Culture A | Culture B | ||
| 1 | 2 | Str | Nal and Str |
| 2 | 2 | Str | Nal and Str |
| 3 | 2 | Str | Str |
| 4 | 4 | Nal | Str |
| 5 | 4 | Nal | Nal |
| 6 | 6 | Nal | Nal and Str |
| 7 | 6 | Str | Str |
| 8 | 8 | Str | Str |
| 9 | 10 | Nal and Str | Nal |
| 10 | 16 | Nal | Nal |
See Fig. 2A for description of the culture pair.
Divergence was observed after the week indicated.
Limit of direction is <102 cfu ml−1.
Quite surprisingly, however, the patterns of fluctuation of NalR and StrR subpopulations remained parallel in some cultures for up to 10 weeks (see Fig. 2D.) In one case, the patterns of takeover remained parallel for almost 4 months (Table 1). The simplest interpretation of long-term parallelism is that the mutant cells that took over at later times were present on initial mixing (week = 0) and were able to persist in the culture until conditions arose that allowed the expression of their GASP phenotype. This implies that the culture environments in each pair of cultures remained similar. A very dramatic illustration of this ability to persist is shown in Fig. 2D. StrR cells remained at titers of ≈5 × 105 cfu ml−1 until week 4. Two weeks later, their levels were below the level of detection (<102 cfu ml−1), only to reappear after another 2 weeks at ≈5 × 104 cfu ml−1. This pattern of disappearance and reappearance was identical in both cultures of the pair. We also confirmed that the reappearing strain had not become resistant to the second drug.
An alternative explanation for the observed long-term parallelism is that the particular mutant alleles conferring the GASP phenotype appeared after mixing and were selected in response to a particular environment. This explanation would require that any changes in the culture environments must happen identically in both cultures, and mutants with identical phenotypes would have to be selected simultaneously. As shown in Fig. 2D, the titers of the StrR cells decreased to less than 102 cfu ml−1 and then, over 2 weeks time, increase at least 500-fold. It seems highly unlikely that two mutations could arise independently and simultaneously in separate subpopulations of such small size (fewer than 500 cells in the entire culture).
Although the patterns of subpopulation fluctuation were parallel in all 10 pairs of cultures for some period of time, eventually these patterns diverged (Table 1; Fig. 2 C and D.) This suggests that over time new mutations arise allowing individual cells in these cultures to evolve along different paths. This divergence is expected because, as the mixed populations age, the chance of the same mutation arising in both cultures of each pair is very low. Cells within each pair of mixed cultures will acquire different GASP-conferring mutations and the relative numbers of NalR and StrR cells within each pair of cultures will eventually diverge. Divergence was observed in all 10 pairs of cultures, although it occurred at different times (Table 1). After 24 weeks of continuous incubation, seven of the cultures contained only NalR cells, nine had only StrR cells, and four cultures still contained both NalR and StrR cells (Table 1).
Initially Isogenic Populations Evolve Along Different Paths.
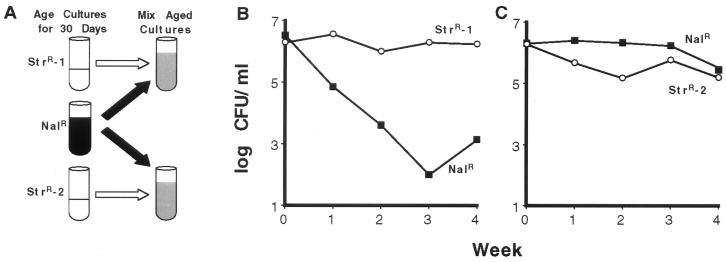
The fact that each of the 10 pairs behaved differently suggests that a high degree of genetic diversity was present in these aged cultures at the time of mixing, which allowed cells to evolve along different paths in waves of takeovers by different GASP mutants. We designed another mixing experiment to directly determine whether initially isogenic bacteria can evolve along different paths when placed in initially identical environments. In these experiments, two StrR cultures (StrR-1 and StrR-2; derived from the same initial culture) and a single NalR culture were aged for 30 days (Fig. 3A). At that time (which becomes week 0 in the experiment) cells from the aged NalR culture were mixed separately with cells from either the StrR-1 or StrR-2 culture, incubation was continued, and their relative titers were determined weekly. As shown in Fig. 3 B and C, cells from the NalR culture are quickly outcompeted by cells from culture StrR-1, but are good competitors against cells from culture StrR-2. This indicates that even though the same culture was used to initially inoculate the two StrR cultures, during the 30 days of separate incubation different mutations were selected that gave cells in each new culture a distinct competitive advantage.
Figure 3.
Initially isogenic strains evolve along different paths. (A) One NalR and two StrR cultures were incubated for 30 days. The NalR culture was mixed 1:1 with cells from either the StrR-1 or StrR-2 cultures, creating two new mixed cultures. The two StrR cultures (StrR-1 and StrR-2) were derived from the same colony and were initially isogenic. (B and C) Representative pair of mixed cultures. NalR cells (■), StrR cells (○).
Different Subpopulations Coexist During Long-Term Incubation.
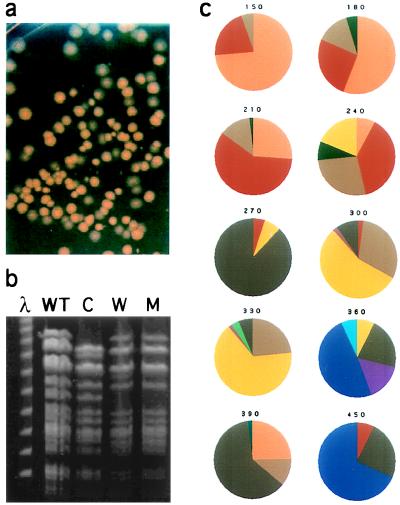
Although we have observed that after the first 10 days of aging of bacterial cultures GASP takeovers seem complete, later takeovers are not complete, as shown in Fig. 1C. This suggests that on continued incubation different mutants may coexist. This diversity can be directly observed on plating of bacterial cells from an aging culture onto nutrient agar plates. As shown in Fig. 4A, cells plated from a 150-day-old culture fell into three distinct colony morphology classes (morphotypes): colonies of normal “cream” color (C), white colonies (W), and small or “mini” colonies (M). The colony morphologies of these cells are likely conferred by genetic factors, because they are stably inherited; on restreaking of a particular colony, the distinctive morphologies breed true. In addition, restriction fragment length polymorphism analysis demonstrates that these distinctive colony morphotypes are derived from the same parental strain (Fig. 4B). However, whereas the SfiI digest patterns of these strains are identical for nearly every band, a few differences were observed. For example, the SfiI digest showed that the C strain chromosome has an ≈50 kb deletion (located at ≈93 min), which causes the loss of two restriction fragments and the appearance of a new band (note the doublet in the top band of lane C in Fig. 4B); differences that distinguish the W and M morphotypes were also observed. Digests with NotI and XbaI revealed further genetic differences between the three colony morphotypes (data not shown). The appearance and disappearance of distinct colony morphotypes over time within a single culture directly demonstrates both that these aged cultures are dynamic and that over time GASP mutants coexist. On day 150 in this culture the composition was ≈75% C, ≈20% W, and ≈5% M (Fig. 4C). However, 2 weeks later the C morphotype appeared to almost completely take over the culture, only to decrease to about 50% of the population at day 180, and to less than 10% by day 240. Also, over this period two new morphotypes, “fried egg” (E) and very small or “nano” (N), appeared. Fluctuations in the proportion and class of colony morphotypes continued to be observed over many months in this particular culture (Fig. 4C), as well as in other initially isogenic cultures (data not shown). These experiments conclusively demonstrate that populations in these long-term cultures are continually dynamic and that takeovers in these environments are rarely complete, giving rise to the evolution of diversity along many different paths.
Figure 4.
Coexistence of different colony morphotypes during long-term incubation in a single culture. (A) Colonies plated from a 150-day-old culture: normal or cream-colored colony morphotype (C), white morphotype (W), and minicolony morphotype (M). (B) Pulsed-field gel electrophoresis of SfiI digested chromosome of wild-type (WT) cells and the three new morphotypes (C, W, and M); a phage lambda (λ) size standard is at left. (C) Pie charts reflecting population composition (%) during different times of incubation (days) of each morphotype, each color represents a different colony morphotype (see text for descriptions).
DISCUSSION
The consecutive takeovers we observe can be thought of as similar to the periodic selections described in chemostats and for long-term serially passaged cultures (5, 13), except that the selections described here arose during long-term stationary phase incubation. The rapidity of takeover and the diversity generated in our system is striking, especially given the small population sizes. Long-term cultures that have been aged for as little as an additional week invariably contain cells that have a competitive advantage over cells from younger cultures. In addition, it is clear that there are many different mutations that can confer a GASP phenotype on these aging cultures. What are the possible mechanisms by which this diversity of mutant alleles is generated?
The starvation conditions encountered during stationary phase incubation may permit a transient increase in the mutation rate due to a variety of factors, including decreased fidelity during replication and reductions in repair activity (14–18). Although an overall increase in mutation frequency might be thought of as being deleterious during times of nutrient deprivation, several models of modulation of mutation frequency during stationary phase (18), as well as experimental evidence (14–17), suggest that in the face of extremely limiting nutrient resources and intense competition, an increase in mutation frequency is a potential mechanism to quickly generate new alleles. In addition, evidence that stationary phase cells may have more than one chromosome equivalent suggests that a transient increase in mutation frequency might not be so problematic (19). With several chromosome equivalents per cell, the effect of deleterious mutations will be masked by other copies of the mutated gene, whereas advantageous mutations will allow proliferation of cells that retain the beneficial allele. Two likely and not mutually exclusive mechanisms responsible for generating cells with more than one chromosome equivalent are (i) an uncoupling of the replication machinery from the cell division apparatus, such that all rounds of replication are completed once initiated, whether or not the cell has the ability to continue dividing (19) and (ii) oriC-independent replication referred to as “inducible stable DNA replication,” where DNA replication is initiated in the absence of cell division (20).
Another source of new alleles may be the dead cells within the culture. For every cell that survives after 30 days of incubation, approximately 1,000 cells have died. Although there is little evidence for natural competence or horizontal gene transfer independent of conjugative mechanisms in E. coli, it is tempting to speculate that cells that find themselves surrounded by the detritus, including chromosomal DNA, of many dead sibling cells, may have an ability to “take up” and incorporate exogenously derived genetic information.
The work presented here makes it clear that as batch cultures age under starvation conditions a wide variety of beneficial mutations arise leading to the evolution of microbial diversity. In their natural setting, bacteria rapidly consume available nutrients and then most likely spend much of their existence in a starved state (21). When nutrients are abundant, selective pressures to increase genetic variation are low. However, upon onset of starvation, there is intense selective pressure for any mutation that confers a competitive advantage. An attractive model of bacterial evolution is one in which the bulk of new mutations is generated during periods of starvation. We would like to propose that our studies of bacterial populations evolving in the laboratory may reflect evolutionary processes acting on natural bacterial populations. When microbes occupy a new nutritional niche they can quickly consume available nutrients and, in all likelihood, enter a state of starvation akin to stationary phase. In these nutritionally poor environments, those bacteria able to scavenge nutrients trapped in the dead biomass will have a selective growth advantage. Over time, as the selective advantage conferred by new advantageous mutant alleles is less pronounced, different GASP mutants can coexist, eventually leading to the evolution of microbial diversity.
Acknowledgments
We thank members of the Kolter and Beckwith labs; Stephen Jay Gould, Michael Farrell, and Linc Sonenshein for insightful comments; and William Rosche and Patricia Foster for helpful discussions and assistance with pulsed-field gel electrophoresis. This work was supported by the National Science Foundation (MCB-9728736) and the National Institutes of Health (GM55199). S.E.F. is a fellow of the Helen Hay Whitney Foundation.
ABBREVIATIONS
- LB
Luria–Bertani
- GASP
growth advantage in stationary phase
- cfu
colony forming units
References
- 1.Novick A, Szilard L. Proc Natl Acad Sci USA. 1950;36:708–719. doi: 10.1073/pnas.36.12.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helling R B, Vargas C N, Adams J. Genetics. 1987;116:349–358. doi: 10.1093/genetics/116.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenski R E, Travisano M. Proc Natl Acad Sci USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elena S E, Lenski R E. Evolution. 1997;51:1058–1067. doi: 10.1111/j.1558-5646.1997.tb03953.x. [DOI] [PubMed] [Google Scholar]
- 5.Dykhuizen D E. Annu Rev Ecol Syst. 1990;21:373–398. [Google Scholar]
- 6.Atwood K C, Schneider L K, Ryan F J. Proc Natl Acad Sci USA. 1951;37:146–155. doi: 10.1073/pnas.37.3.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zambrano M M, Siegele D A, Almirón M, Tormo A, Kolter R. Science. 1993;259:1757–1759. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 8.Zambrano M M, Kolter R. J Bacteriol. 1993;175:5642–5647. doi: 10.1128/jb.175.17.5642-5647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zambrano M M, Kolter R. Cell. 1996;86:181–184. doi: 10.1016/s0092-8674(00)80089-6. [DOI] [PubMed] [Google Scholar]
- 10.Finkel S E, Zinser E, Gupta S, Kolter R. In: Molecular Microbiology: NATO-ASI Series. Busby S J W, Thomas C M, Brown N L, editors. Berlin: Springer; 1997. pp. 3–16. [Google Scholar]
- 11.Miller J H. A Short Course in Bacterial Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 12.Koob M, Szybalski W. Methods Enzymol. 1992;216:13–20. doi: 10.1016/0076-6879(92)16004-4. [DOI] [PubMed] [Google Scholar]
- 13.Elena S F, Cooper V S, Lenski R E. Science. 1996;272:1802–1804. doi: 10.1126/science.272.5269.1802. [DOI] [PubMed] [Google Scholar]
- 14.Foster P L. J Bacteriol. 1997;179:1550–1554. doi: 10.1128/jb.179.5.1550-1554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torkelson J, Harris R S, Lombardo M J, Thulin C, Rosenberg S M. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bridges B A. Nature (London) 1997;387:557–558. doi: 10.1038/42370. [DOI] [PubMed] [Google Scholar]
- 17.Sniegowski P D, Gerrish P J, Lenski R E. Nature (London) 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 18.Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon P H, Godelle B. Nature (London) 1997;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- 19.Åkerlund T, Nordström K, Bernander R. J Bacteriol. 1995;177:6791–6797. doi: 10.1128/jb.177.23.6791-6797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kogoma T. Microbiol Mol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morita R M. In: Starvation in Bacteria. Kjellberg S, editor. New York: Plenum; 1993. pp. 1–23. [Google Scholar]