Abstract
The rugose colony variant of Vibrio cholerae O1, biotype El Tor, is shown to produce an exopolysaccharide, EPSETr, that confers chlorine resistance and biofilm-forming capacity. EPSETr production requires a chromosomal locus, vps, that contains sequences homologous to carbohydrate biosynthesis genes of other bacterial species. Mutations within this locus yield chlorine-sensitive, smooth colony variants that are biofilm deficient. The biofilm-forming properties of EPSETr may enable the survival of V. cholerae O1 within environmental aquatic habitats between outbreaks of human disease.
The epidemiology of cholera in the Bengal region of India and Bangladesh is marked by periodic, seasonal outbreaks followed by long intervals during which the disease occurs sporadically or not at all (1). Failure to regularly identify chronic carriers of Vibrio cholerae O1 or infected animal reservoirs, its capacity to colonize the gut of copepods (2), and detection of the organism attached to phytoplankton (3–5) and in water samples throughout the year (6) has led to the idea that V. cholerae O1 resides within natural aquatic habitats during inter-epidemic periods (7). Possibilities suggested by other investigators include its persistence in a viable, but nonculturable, state in water (8, 9) and its association as a commensal or symbiont of other members of the aquatic flora (7). These microenvironments have in common the survival of the organism under physiological constraints that likely differ markedly from conditions within the human gastrointestinal tract. A common feature of most environmental reservoirs is the low availability of nutrients, compared with the intestinal milieu. Additionally, environmental habitats are subject to seasonally determined changes of the microflora and to physicochemical fluctuations. Here we report the following: the rugose colonial variant of V. cholerae O1, biotype El Tor produces a unique extracellular polysaccharide, designated EPSETr, that confers resistance to chlorine and promotes biofilm formation. Compositional and linkage analysis of this material shows it to be unrelated to previously described V. cholerae carbohydrates, and mutational experiments led to the identification of a chromosomal cluster of genes that is required for the production of this compound and for rugose-associated phenotypes. In view of the functional properties it confers, we now propose a role for EPSETr in the survival of the organism within environmental aquatic habitats.
MATERIALS AND METHODS
Bacterial Strains.
Escherichia coli strains DH5α and S17–1 (10) were used for standard DNA manipulations and mating, respectively. The V. cholerae strains used were smooth and rugose variants of 92A1552 (wild type, El Tor, Inaba, and Rifr) and mutants of these strains listed in Table 2.
Table 2.
Sequence analysis of transposon-tagged genes reveals homologies with polysaccharide biosynthesis motifs
| Mutant strain | Homologous protein | Degree of homology, % identity/ % similarity | Function of the homologous protein | Organism |
|---|---|---|---|---|
| 38, (2*, 44*, 49*) | Ipi1 | 25.0/33.8 | Isopentenyl-diphosphate delta-isomerase | Arabidopsis thaliana |
| 3 | YwlE | 37.9/51.7 | Protein tyrosine phosphatases | Bacillus subtilis |
| 65, (8, 41) | ExoP | 22.3/35.2 | Polymerization and/or export of succinoglycan | Rhizobium meliloti |
| 21, (24) | EpsD | 74.5/80.1 | NDP-N-acetyl-d-galactosaminuronic acid dehydrogenase | Ralstonia solanacearum |
| 32, (23, 60) | AmsK | 34.1/43.3 | Biosynthesis of amylovoran | Erwinia amylovora |
| 59 (25*) | CysE | 17.0/21.9 | Serine acetyl transferase | Synechocystis PCCC6803 |
| 54, (31, 35, 68) | GumD | 45.0/38.4 | Glycosyl-1-phosphate transferase | Xanthomonas campestris |
| 39 | EpsC | 66.2/69.8 | UDP-N-acetylglucosamine 2-epimerase | R. solanacearum |
| 50 | Cap1K | 16.2/26.5 | UDP-glucose dehydrogenase | Streptococcus pneumoniae |
| 69 | ExoF | 28.9/36.1 | Biosynthesis of succinoglycan | R. meliloti |
| 70 | CapK | 28.9/33.3 | Biosynthesis of type1 capsular polysacchardie | Staphylococcus aureus |
| 74 | ORF11 | 25.8/36.6 | Biosynthesis of serotype K2 capsular polysaccharide | Klebsiella pneumoniae |
| 76 | WcaL | 17.9/28.0 | Glycosyl transferase | Escherichia coli |
| 4, 9, 18, 73 | None |
Based on the identification of homologous sequences of proteins of known function. Degree of homology represented in the third column corresponds to the numbers shown in bold, in the first column.
Homologies were determined after identification of overlapping contigs from the V. cholerae O1 tigr sequencing project.
Microscopy.
Scanning and transmission electron microscopy, ruthenium red staining, and immunogold electron microscopy were performed as described (11).
Isolation of EPS.
Smooth or rugose colonies were cultivated for 24 hr at 30°C on the surface of sterile cellophane dialysis membranes placed on the surfaces of LB agar plates. The lawn of confluent bacterial growth was harvested and suspended in 0.9% NaCl. EPSETr then was detached from the bacterial surface and purified according to published methods (12–14). Glycosyl composition and linkage analysis was performed at the University of Georgia’s Complex Carbohydrate Research Center (15).
Mutagenesis.
Transposon mutagenesis of V. cholerae O1 El Tor, strain 92A1552, was performed by conjugation with the donor E. coli S-17-l λpir, containing a pool of ≈40,000 signature-tagged pUT mini-Tn5-Km2 (16). The exconjugants were selected by plating the suspension onto LB plates supplemented with 100 μg/ml of rifampicin and 150 μg/ml of kanamycin.
DNA Manipulations and Analysis.
Plasmid DNA and chromosomal DNA preparation, DNA ligation, bacterial transformation, agarose gel electrophoresis, Southern blotting, and cosmid library construction were performed by standard methods (17).
Gene Cloning and DNA Sequencing.
Chromosomal DNA was singly digested with EcoRI, SalI, KpnI, and PstI, and the fragments were resolved on an agarose gel, transferred to Hybond N membranes, and hybridized to the kanamycin-resistance gene of pUT mini-Tn5-Km2. Restriction enzymes that yielded hybridizing fragments between 3 and 8 kb were used to digest chromosomal DNA, and the resulting fragments were cloned into BluescriptKS. Plasmids were analyzed by restriction enzyme digestion, and a region of ≈300 bp was sequenced from the region flanking the transposon insertion site. The sequence was analyzed by the gap program of the GCG software package and blast of the National Center for Biotechnology Information over a minimum 50-aa sequence.
Chlorine Killing, Biofilm, and EPS Assays.
For chlorine survival analysis, 1–5 × 107 bacteria were incubated with 3 ppm NaOCl for 0 min and 5 min at 30°C. Surviving bacteria were enumerated at the two time points by viable plate counts of bacteria that had been vortexed with 3-mm glass beads to form a suspension of single cells. Biofilm formation was assayed as described (18). EPS production was detected by using a solid-phase assay of culture supernatants and an EPS-specific antiserum.
RESULTS AND DISCUSSION
The Rugose Colony Variant of V. cholerae 01 El Tor.
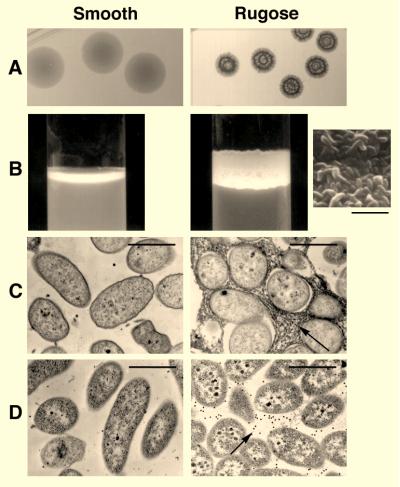
V. choleare O1 Inaba, biotype El Tor, strain 1552, isolated in 1992 from a patient with cholera who had traveled recently in Latin America, was cultivated at 30°C with aeration for 20 days under conditions of carbon limitation in M9 minimal medium supplemented with 0.02% glucose. Plating of the starting (carbon-replete) and final (carbon-starved) cultures onto LB agar showed that a striking change in colonial morphology had occurred: ≈ 1% of the colonies grown from the starved culture were wrinkled and opaque (Fig. 1A) compared with the smooth and translucent colonies of the unstarved culture. In static nutrient broth, these wrinkled colonies also were noted to form floating pellicles containing closely packed bacteria (Fig. 1B Inset). By contrast, the smooth colonial variant grew below the surface of the broth as a suspension of single cells.
Figure 1.
The rugose colonial variant of V. cholerae O1 El Tor produces an extracellular glycocalyx. (A) Colonial morphology of the smooth and rugose variants grown on LB agar at 30°C for 72 hr. (B) Pellicle formation by the rugose variant grown in LB broth for 72 hr. The rugose colony type forms a floating membrane at the air-broth interface where it coats the glass surface of the culture tube. Scanning electron microscopy of the pellicle shows it to be composed of closely packed bacteria (Inset). The smooth variant grows mainly below the surface. (C) Ruthenium red-stained thin sections of smooth and rugose colonies. Electron micrographs demonstrate a stained matrix between rugose-type bacteria. (D) Immunogold electron microscopy of thin-sectioned smooth and rugose colonies. An EPSETr-specific antiserum localized the polysaccharide antigen between rugose-type bacteria. [Bars = 5 μm (B) and 1 μm (C and D).]
A review of the older literature disclosed that the wrinkled colony type was in fact the rugose variant described by Balteanu in 1926 and White in 1938 (19). More recently Rice et al. (20) isolated rugose variants during the recent cholera epidemic in Latin America and showed that rugosity is associated with increased survival in chlorinated water, when compared with the smooth-colony form, and Morris and colleagues (21) showed that the rugose variant could cause a typical cholera diarrheal illness when orally administered to volunteers, an observation indicating that the rugose form is not merely a laboratory curiosity. A recent study of the rugose variant by Wai et al. (11) showed that it could produce a capsule-like surface layer and attach better to glass than the smooth variant (11).
To learn whether in vitro switching between the smooth and rugose colony types occurs during cultivation in nutrient broth, single colonies of the smooth or rugose variant were inoculated into separate flasks of LB broth and then incubated for 5 days at 30°C with aeration. At 24 hr and 120 hr serial dilutions of the cultures were vortexed with 3-mm glass beads to produce single-cell suspensions and inoculated onto LB agar, and the number of rugose and smooth colonies were counted. By 24 hr of incubation, 0.05% of the smooth-type bacteria had converted to the rugose morphotype and by day 5 the proportion of rugose-type bacteria had increased to 6.0%. Switching from the rugose to the smooth morphotype also was observed: by day 5 10.2% of rugose-type bacteria had converted to the smooth-colony form. Thus bidirectional switching between the two colony types occurs and would favor outgrowth of the variant best adapted to a particular environment.
Rugosity Is Associated with EPS Production.
To better appreciate the nature of interbacterial adhesions between rugose-type bacteria, we grew each colony type on separate sterile cellophane membranes placed on the surface of LB agar and then examined the resulting bacterial film by staining ultrathin sections with ruthenium red followed by transmission electron microscopy. This method, which preferentially stains acidic polysaccharides (22), revealed an abundant, ruthenium red-positive matrix between rugose-type bacteria (Fig. 1C); no such material was evident in films composed of the smooth colonial variant (Fig. 1C). In contrast, the recently described capsule-like layer of a rugose variant of the El Tor biotype by Wai and colleagues (11) was found to be closely associated with the bacterial surface.
The ruthenium red-positive material was isolated and subjected to glycosyl composition and linkage analysis (15) to determine whether it was identical to previously characterized V. cholerae polysaccharides. It was found to contain nearly equal amounts of glucose and galactose, with smaller amounts of N-acetylglucosamine and mannose (Table 1). This compositional result clearly distinguishes it from the capsule-like material isolated by Wai et al.(11) from a rugose variant of a V. cholerae El Tor strain, which they reported lacked glucose and contained 6-deoxy-d-galactose; from V. cholerae O1 lipopolysaccharide, which contains large amounts of perosamine and quinovosamine (23–25); and from the capsular polysaccharide of V. cholerae O139, which apparently evolved from an El Tor progenitor (26, 27) and contains 3,6-dideoxyxylohexose (28). Linkage analysis detected nearly equal amounts of 4-linked galactose and 4-linked glucose (Table 1) and these therefore may comprise the backbone(s) of the saccharide. Branching is indicated by the presence of 3,4- and 4,6-linked galactose and glucose and by 2,4-linked galactose (Table 1). Further work will disclose the complete structure and whether it contains acetate, pyruvate, or succinate—common substituents of bacterial exopolysaccharides that would increase the hydrophobicity of the compound and, when ketal-linked, would impart a negative charge (29, 30).
Table 1.
Glycosyl composition and linkage analysis of EPS
| Glycosyl composition
|
Glycosyl linkage
|
||
|---|---|---|---|
| Glycosyl residue | Percent* | Glycosyl residue | Relative ratio† |
| Glucose | 52.6 | Terminal glucose | 0.19 |
| Galactose | 37.0 | Terminal galactose | 0.06 |
| GlcNAc | 5.1 | 3-linked galactose | 0.21 |
| Mannose | 3.8 | 4-linked galactose | 0.89 |
| Xylose | 1.5 | 4-linked glucose | 1.0 |
| 3,4-linked galactose | 0.19 | ||
| 3,4-linked glucose | 0.17 | ||
| 2,4-linked galactose | 0.17 | ||
| 4,6-linked glucose | 0.11 | ||
| 4,6-linked galactose | 0.09 | ||
Values are expressed as the weight percent of total carbohydrate.
Values represent the ratios of the GC peak areas with terminal 4-linked glucose set to 1.0.
To determine whether this material was localized to the same interbacterial compartment as the ruthenium red-positive material shown in Fig. 1C, an antiserum to rugose-type bacteria was elicited in a rabbit, absorbed with smooth-type bacteria and used to conduct immunogold electron microscopy of thin-sectioned smooth and rugose colonies. Gold particles localized between adjacent rugose-type bacteria, but did not bind between or within smooth-type bacteria (Fig. 1D). As a result of these findings we propose the designation EPSETr, to denominate the extracellular polysaccharide produced by the rugose variant of the El Tor biotype.
EPSETr Mediates Chlorine Survival and Biofilm Formation.
Chlorine has been the principal means of preventing waterborne infectious diseases, including cholera, since the first decade of this century (31). To learn whether EPSETr might be responsible for the relative chlorine resistance of the rugose variant reported by others (20), we carried out survival experiments of the smooth variant in sodium hypochlorite (NaOCl) containing different concentrations of EPSETr. For the experiments described below, we used concentrations of NaOCl that are 10- to 20-fold higher than the free chlorine concentrations typically obtained in municipal water supplies.
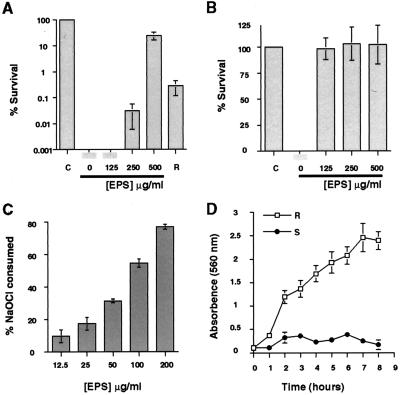
NaOCl (3 ppm) was incubated with 2.0 × 107 colony-forming units of smooth-type bacteria, and the surviving bacteria were enumerated by plate counts. No smooth-type bacterial survivors were detected after a 5-min exposure period (Fig. 2A). By contrast, under the same experimental conditions, 6 × 104 rugose-type bacteria, as determined by plate counts of dispersed bacteria, survived NaOCl exposure, confirming the observation of Rice et al. (20) that the smooth variant is chlorine sensitive, whereas the rugose variant is relatively chlorine resistant. However, smooth-type bacteria could be converted to a state of relative chlorine resistance by the addition of EPSETr: the presence of 125 μg/ml, 250 μg/ml, and 500 μg/ml of EPSETr resulted in 0%, 0.03% and 25% survival, respectively, of smooth-type bacteria exposed to 3 ppm NaOCl for 5 min. This protective effect could be dramatically increased by preincubation of NaOCl with EPSETr before the addition of these reactants to a suspension of smooth-type bacteria. Under this experimental condition, the lowest tested concentration of EPSETr (125 μg/ml) conferred complete protection (Fig. 2B) against 3 ppm NaOCl. Taken together, these experiments suggest that EPSETr is directly responsible for the chlorine resistance of the rugose variant. This possibility was further tested by determining whether EPSETr could consume or otherwise inactivate chlorine, a property previously shown for the alginic acid-containing slime produced by mucoid variants of Pseudomonas aeruginosa (32). NaOCl (6 ppm) was incubated for 1 min with different concentrations of EPSETr; the resulting free chlorine concentration then was measured by the syringaldazine method (33). In the absence of EPSETr no change in chlorine concentration was observed. By contrast, in the presence of EPSETr, a dose-dependent consumption of free chlorine was detected (Fig. 2C); at the highest tested concentration of EPSETr (200 μg/ml), 80% of the starting concentration of free chlorine was consumed within the 1-min reaction period.
Figure 2.
EPSETr-mediated survival in chlorine and biofilm formation by the rugose and smooth colonial variants. (A) Smooth-type bacteria were incubated for 5 min with 3 ppm chlorine (NaOCl) and the indicated concentrations of purified EPSETr. The surviving bacteria were enumerated by viable plate counts, and their numbers were compared with the number of surviving smooth bacteria that had not been incubated with NaOCl, denoted by C; this number was defined as 100% survival. Increasing survival of the smooth variant in chlorine was conferred by EPSETr concentrations of 250 and 500 μg/ml. Rugose and smooth bacteria, incubated with 3 ppm NaOCl in the absence of exogenous EPSETr, denoted by R and 0, respectively, revealed differences between the intrinsic resistances of the two colonial variants to the bactericidal activity of chlorine. (B) Preincubation of NaOCl (3 ppm) with the indicated concentrations of EPSETr for 5 min before the addition of smooth-type bacteria increased the protective effect of the polysaccharide. C denotes the survival of smooth-type bacteria in the absence of NaOCl and EPSETr. (C) Consumption of free chlorine by purified EPSETr. NaOCl (6 ppm) and the indicated concentrations of EPSETr were incubated in PBS for 1 min at 22°C, and the free chlorine concentration then was determined by the syringaldazine method. EPSETr caused rapid, concentration-dependent consumption of chlorine. (D) Quantitative analysis of biofilm formation by the smooth and rugose variants. Bacteria of each colony type (1–5 × 106 colony-forming units) were incubated in separate wells of a poly(vinyl chloride) microtiter dish for the indicated time periods, the wells were emptied and washed, and the adherent bacteria were stained with a 1% solution of crystal violet. The dye was solubilized by the addition of 95% ethanol, and the absorbance was determined at 560 nm. Progressive biofilm formation by the rugose variant is indicated by an increase in A560 during the experimental period.
The propensity of the rugose colonial variant to coat the glass side wall of a culture tube, evident in Fig. 1B, prompted us to formally study its biofilm-forming behavior. Each colonial variant was grown in separate wells of nine poly(vinyl chloride) microtiter plates, and the biofilm was quantified at time 0 and once each hour thereafter. Rugose-type bacteria showed a time-dependent increase in surface colonization (Fig. 2D). By contrast, the smooth colonial variant adhered poorly during the entire 8-hr observation period. To learn whether the apparent differences in biofilm forming capacity between the two colonial variants might have been caused by differences in their growth rates, they were cultivated for 8 hr in the same medium, and their concentrations were determined each hour by nephelometry after being dispersed as single-cell suspensions. No differences in growth rate were detected (data not shown).
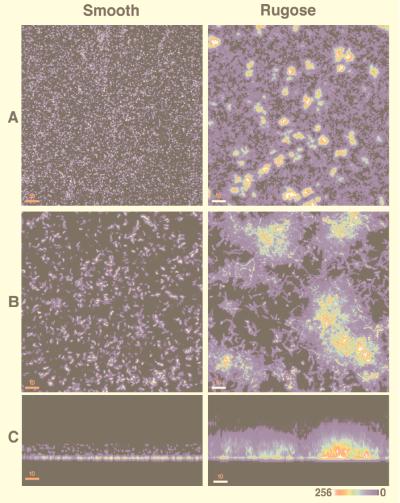
To learn more about the three-dimensional architecture of the adherent bacteria, we conducted scanning confocal laser microscopy (SCLM) using smooth and rugose bacteria carrying a plasmid-encoded locus that constituitively expresses the green fluorescent protein (34). Smooth and rugose bacteria were separately incubated in chambers containing borosilicate coverglasses. After 1, 2, 4, 6, and 24 hr, the wells were emptied and examined by SCLM. Optical sections from the substratum to the biofilm surface were obtained. Serial horizontal (xy) sections were projected by using a maximum-intensity algorithm and, when viewed at low magnification, showed that the rugose variant forms discrete, irregular islands of surface-adherent bacterial aggregates within 1 hr of incubation. In comparison, smooth-type bacteria attached to the substratum as individual cells (Fig. 3 A and B). By hour 6, approximately 4% of the substratum was composed of aggregate-associated bacteria; between aggregates, individual bacteria were seen adhering to the glass surface (Fig. 3B). Reconstruction of these images along the xz axis yielded a sagittal profile of the biofilm and showed that the aforementioned aggregates formed peaks and ridges that averaged 30.3 μm in height by the 6-hr time point (Fig. 3C). Little change in the height of the adherent aggregates occurred with aging of the biofilm in this static system. By contrast, smooth-type bacteria formed a low-profile biofilm, averaging 11.7 μm in height (Fig. 3C).
Figure 3.
Three-dimensional reconstructions of biofilms formed by the smooth and rugose colonial variants. Smooth and rugose type bacteria carrying a plasmid constituitively expressing the green fluorescent protein were incubated in chambers containing borosilicate coverglass bottoms. The wells were emptied at 1, 2, 4, 6, and 24 hr, washed, and examined with a scanning confocal laser microscope (MultiProbe 2010, Molecular Dynamics) using 488- and 510-nm excitation and emission wavelengths, respectively. (A and B) Horizontal (xy) projected images at low and high magnification, respectively, of biofilms formed by the two colony types after a 6-hr incubation period. Islands of adherent bacterial aggregates typify the rugose-type biofilm. (C) Image reconstruction led to a saggital (xz) view of the same biofilms and revealed dramatic differences between their heights. The relative intensity of the pseudo-colored images is shown at the lower right corner and correlates with cell density. [Bars 50 μM (A) and 10 μM (B and C)] ×18.
Identification of a Gene Cluster Required for Phenotypes of the Rugose Colony Variant.
The traits thus far associated with the rugose phenotype—–a distinctive colonial morphology, production of EPSETr, chlorine resistance, and biofilm formation—–could be coincidentally related or coregulated or determined by gene(s) at a single locus. To address this question, the rugose colonial variant was subjected to transposon mutagenesis, and the resulting transconjugants were screened by low-power microscopy to identify mutants that had undergone a rugose-to-smooth transition in colonial morphology. Of 150,000 transconjugants screened, we analyzed 29 that were phenotypically smooth. Unlike the wild-type smooth variant, none of these mutants reverted to the rugose phenotype. The transposon insertion sites were cloned by marker rescue, and the flanking regions were sequenced. Six of the sequences were found in more than one clone, indicating that transposon insertion had reached saturation. The protein sequences deduced from 20 of these sequence tags were homologous to 11 functionally different EPS and capsular biosynthesis proteins from other species. Included were proteins with the following functions: nucleotide sugar precursor synthesis; glycosyl transferases and polymerases; EPS secretion; and EPS modification by the addition of noncarbohydrate groups (35–43) (Table 2). An additional sequence tag corresponded to homologues of tyrosine phosphatase genes found in the EPS gene clusters of Ralstonia solanacearum and Erwinia amylovora (38) (Table 2). Of the 29 mutants, only four had no identifiable homologue and none were part of the V. cholerae serotype O1 lipopolysaccharide or O139 capsule gene clusters (44, 45).
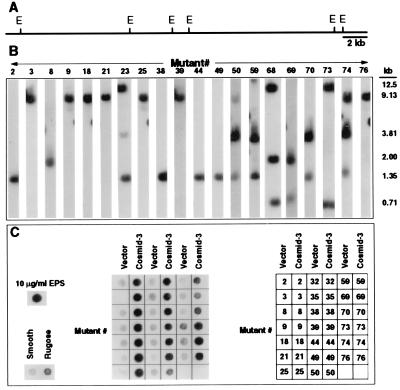
To determine whether these sequences were physically linked on the V. cholerae O1 chromosome, a cosmid library was prepared from the smooth variant and overlapping cosmids containing EPS sequences that were identified by screening the library using as probes the sequence tags from mutants 8, 21, 35, and 74 (Table 2). Restriction and Southern analysis (Fig. 4 A and B) showed that all 29 sequences mapped to cosmid-3, which spans 30.7 kb on the V. cholerae chromosome. Twenty of the transposon mutants, representing each of the 14 homology groups listed in Table 2, were analyzed for EPSETr production by using the antiserum described above and in Fig. 1D. All were EPSETr antigen-negative (Fig. 4C). However, the rugose morphotype and EPSETr production were restored in each by complementation with cosmid-3 (Fig. 4C). Six of the mutants (nos. 21, 35, 39, 50, 59, and 69, Table 2 and Fig. 4), harboring either the cloning vector or cosmid-3, also were tested for chlorine sensitivity and biofilm-forming capacity. All of the tested mutants were completely killed when incubated with 3 ppm NaOCl for 5 min at 30°C, whereas each of the mutants complemented by cosmid-3 had rugose type levels of chlorine resistance. Similarly, these mutants were unable to form rugose-type biofilms, but they recovered biofilm-forming capacity when complemented with cosmid-3 (data not shown). Cosmid-3 therefore encompasses a gene cluster required for EPSETr production, which we designate vps (Vibrio polysaccharide synthesis). These results also provide compelling evidence that vps and its product, EPSETr, are required for each of the rugose-associated traits described above.
Figure 4.
Identification of sequences on the V. cholerae O1, El Tor chromosome that are required for EPSETr production. (A) Physical map of cosmid-3 derived from restriction mapping and Southern analysis of insert DNA. E denotes EcoRI restriction sites. (B) Location of tagged DNA fragments within cosmid-3. Cosmid-3 was digested with EcoRI, the restriction fragments were separated by 0.8% agarose gel electrophoresis, transferred onto membranes, and hybridized with radiolabeled sequence tags from each of the indicated mutants (identified by the number above each autoradiograph and corresponding to the numbered designation in Table 2). The sizes of the hybridized fragments are indicated. (C) EPSETr production by the mutants harboring either the cloning vector alone or complemented with cosmid-3, as determined by ELISA on a nitrocellulose membrane using an EPSETr-specific antiserum. Supernatants of the smooth (S) and rugose (R) variants containing crude EPSETr were included as negative and positive controls, respectively. Purified EPSETr was used as an additional positive control. Cosmid-3 complemented EPSETr production by each of the mutants.
Use of the sequence tags as probes to study the classical biotype of V. cholerae O1 revealed that it also contains the vps gene cluster (data not shown). However, we were unable to induce the classical strains in our collection to adopt the rugose phenotype. EPSETr production by the classical biotype may require different induction conditions. Alternatively, the classical biotype might lack critical regulatory or accessory genes or its vps gene cluster may be incomplete. If so, this could explain the reported superior capacity of the El Tor biotype to persist in environmental reservoirs (46, 47).
The aquatic phase of the V. cholerae O1 life cycle seems to encompass several stages; transitions between these stages are likely controlled by physicochemical features of the habitat, which, in turn, are climate determined. Stage-specific adaptation to changing environmental conditions would account for this organism’s capacity to persist within aquatic habitats as a more or less permanent member of the autochthonous flora (2, 7, 48). With respect to the biofilm stage, a two-compartment model has been suggested (18, 49, 50), with oscillations between free-swimming, individual bacteria and the bacterial population within surface-attached communities. Further complexity is introduced by additional stages involving zooplankton, phytoplankton, and the viable, but nonculturable, state (7, 8).
Our findings that the rugose colonial variant of V. cholerae O1, biotype El Tor expresses an EPS led us to focus on the biofilm stage of the organism’s environmental life cycle. Within the biofilm, bacteria can access trapped and adsorbed nutrients, engage in favorable metabolic transactions with other members of the biofilm (which in nature may contain heterologous species), and be protected from grazing predators (51, 52). The observation that EPSETr confers chlorine resistance predicts that it also may provide protection from natural environmental oxidants and indeed, the rugose variant has been shown to be relatively resistant to H2O2 (11). Emigration from the biofilm could spawn new communities elsewhere, a process that might require localized digestion of the interbacterial matrix. Besides contributing to the release of biofilm organisms, the release phase of the process could provide a utilizable source of carbon. Viewed in this way, the biofilm stage is itself a multistage, multifunctional phenotype.
Our studies were performed with synthetic surfaces and media. Now it will be important to learn whether V. cholerae O1 El Tor can be identified within biofilms in naturally infected habitats and to correlate the biofilm-forming stage with climate change and cholera epidemiology.
Acknowledgments
We thank R. Fernandez for performing the electron microscopy, S. L. Palmieri for performing scanning confocal laser microscopy, R. Valdivia and S. Falkow for providing the green fluorescent protein plasmid, D. Holden for providing the signature tagged mutagenesis pool, and D. Bieber, D. Kaiser and B. Stocker for critical reading of the manuscript. Carbohydrate analysis was conducted by the Complex Carbohydrate Research Center of the University of Georgia. This work was supported by Grant RO1-AI43422 from the National Institutes of Health.
ABBREVIATIONS
- EPS
exopolysaccharide
- vps
Vibrio polysaccharide synthesis
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. (vps38) AF093222, (vps3) AF093223, (vps65) AF093224, (vps21) AF093225, (vps32) AF093226, (vps59) AF093227, (vps54) AF093228, (vps39) AF093229, (vps50) AF093230, (vps69) AF093231, (vps70) AF093232, (vps74) AF093233, (vps76) AF093234, (vps4) AF093235, (vps9) AF093236, (vps18) AF093237, and (vps73) AF093238].
References
- 1.Glass R I, Claeson M, Blake P A, Waldman R J, Pierce N F. Lancet. 1991;338:791–795. doi: 10.1016/0140-6736(91)90673-d. [DOI] [PubMed] [Google Scholar]
- 2.Colwell R R, Spira W M. In: The Ecology of Vibrio cholerae. Barua D, Greenough III W B, editors. New York: Plenum; 1992. pp. 107–123. [Google Scholar]
- 3.Islam M S, Drasar B S, Bradley D J. J Trop Med Hyg. 1990;93:133–139. [PubMed] [Google Scholar]
- 4.Islam M S, Drasar B S, Sack R B. J Diarrhoeal Dis Res. 1994;12:245–256. [PubMed] [Google Scholar]
- 5.Kaper J, Lockman H, Colwell R R, Joseph S W. Appl Environ Microbiol. 1979;37:91–103. doi: 10.1128/aem.37.1.91-103.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huq A, Colwell R R, Rahman R, Ali A, Chowdhury M A, Parveen S, Sack D A, Russek-Cohen E. Appl Environ Microbiol. 1990;56:2370–2373. doi: 10.1128/aem.56.8.2370-2373.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Islam M S, Drasar B S, Sack R B. J Diarrhoeal Dis Res. 1994;12:87–96. [PubMed] [Google Scholar]
- 8.Roszak D B, Colwell R R. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiba T, Hill R T, Straube W L, Colwell R R. Appl Environ Microbiol. 1995;61:2583–2588. doi: 10.1128/aem.61.7.2583-2588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yildiz F H, Schoolnik G K. J Bacteriol. 1998;180:773–784. doi: 10.1128/jb.180.4.773-784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wai S N, Mizunoe Y, Takade A, Kawabata S, Yoshida S. Appl Environ Microbiol. 1998;64:3648–3655. doi: 10.1128/aem.64.10.3648-3655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linker A, Evans L R. J Bacteriol. 1984;159:958–964. doi: 10.1128/jb.159.3.958-964.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vann W F, Liu T Y, Robbins J B. Infect Immun. 1976;13:1654–1662. doi: 10.1128/iai.13.6.1654-1662.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Read R R, Costerton J W. Can J Microbiol. 1987;33:1080–1090. doi: 10.1139/m87-189. [DOI] [PubMed] [Google Scholar]
- 15.Merkle R K, Poppe I. Methods Enzymol. 1994;230:1–15. doi: 10.1016/0076-6879(94)30003-8. [DOI] [PubMed] [Google Scholar]
- 16.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.O’Toole G A, Kolter R. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 19.White P. J Pathol Bacteriol. 1938;46:1–6. [Google Scholar]
- 20.Rice E W, Johnson C H, Clark R M, Fox K R, Reasoner D J, Dunnigan M E, Panigrahi P, Johnson J A, Morris J G. Int J Environ Health Res. 1993;3:89–98. [Google Scholar]
- 21.Morris J G, Jr, Sztein M B, Rice E W, Nataro J P, Losonsky G A, Panigrahi P, Tacket C O, Johnson J A. J Infect Dis. 1996;174:1364–1368. doi: 10.1093/infdis/174.6.1364. [DOI] [PubMed] [Google Scholar]
- 22.Patterson H, Irvin R, Costerton J W, Cheng K J. J Bacteriol. 1975;122:278–287. doi: 10.1128/jb.122.1.278-287.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raziuddin S. Infect Immun. 1980;27:211–215. doi: 10.1128/iai.27.1.211-215.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redmond J W. FEBS Lett. 1975;50:147–149. doi: 10.1016/0014-5793(75)80476-5. [DOI] [PubMed] [Google Scholar]
- 25.Hisatsune K, Hayashi M, Haishima Y, Kondo S. J Gen Microbiol. 1989;135:1901–1907. doi: 10.1099/00221287-135-7-1901. [DOI] [PubMed] [Google Scholar]
- 26.Bik E M, Bunschoten A E, Gouw R D, Mooi F R. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comstock L E, Maneval D, Jr, Panigrahi P, Joseph A, Levine M M, Kaper J B, Morris J G, Jr, Johnson J A. Infect Immun. 1995;63:317–323. doi: 10.1128/iai.63.1.317-323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preston L M, Xu Q, Johnson J A, Joseph A, Maneval D R, Jr, Husain K, Reddy G P, Bush C A, Morris J G., Jr J Bacteriol. 1995;177:835–838. doi: 10.1128/jb.177.3.835-838.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jansson P E, Kenne L, Lindberg B. Carbohydr Res. 1975;45:275–282. doi: 10.1016/s0008-6215(00)85885-1. [DOI] [PubMed] [Google Scholar]
- 30.Melton L D, Mindt L, Rees D A, Sanderson G R. Carbohydr Res. 1976;46:245–257. doi: 10.1016/s0008-6215(00)84296-2. [DOI] [PubMed] [Google Scholar]
- 31.Margolin A B. In: Control of Microorganisms in Source Water and Drinking Water. Hurst C J, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Washington, DC: Am. Soc. Microbiol.; 1997. pp. 195–202. [Google Scholar]
- 32.Learn D B, Brestel E P, Seetharama S. Infect Immun. 1987;55:1813–1818. doi: 10.1128/iai.55.8.1813-1818.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper W J, Sorber C A, Meier E P. J Am Water Works Assoc. 1975;67:34–39. [Google Scholar]
- 34.Valdivia R H, Falkow S. Mol Microbiol. 1996;22:367–378. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- 35.Arakawa Y, Wacharotayankun R, Nagatsuka T, Ito H, Kato N, Ohta M. J Bacteriol. 1995;177:1788–1796. doi: 10.1128/jb.177.7.1788-1796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becker A, Kleickmann A, Keller M, Arnold W, Puhler A. Mol Gen Genet. 1993;241:367–379. doi: 10.1007/BF00284690. [DOI] [PubMed] [Google Scholar]
- 37.Becker A, Ruberg S, Kuster H, Roxlau A A, Keller M, Ivashina T, Cheng H P, Walker G C, Puhler A. J Bacteriol. 1997;179:1375–1384. doi: 10.1128/jb.179.4.1375-1384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang J, Schell M. Mol Microbiol. 1995;16:977–989. doi: 10.1111/j.1365-2958.1995.tb02323.x. [DOI] [PubMed] [Google Scholar]
- 39.Long S, Reed J W, Himawan J, Walker G C. J Bacteriol. 1988;170:4239–4248. doi: 10.1128/jb.170.9.4239-4248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morona J K, Morona R, Paton J C. J Bacteriol. 1997;179:4953–4958. doi: 10.1128/jb.179.15.4953-4958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ouyang S, Lee C Y. Mol Microbiol. 1997;23:473–482. doi: 10.1046/j.1365-2958.1997.d01-1865.x. [DOI] [PubMed] [Google Scholar]
- 42.Reuber T L, Walker G C. Cell. 1993;74:269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- 43.Roberts I S. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 44.Stroeher U H, Jedani K E, Dredge B K, Morona R, Brown M H, Karageorgos L E, Albert M J, Manning P A. Proc Natl Acad Sci USA. 1995;92:10374–10378. doi: 10.1073/pnas.92.22.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stroeher U H, Parasivam G, Dredge B K, Manning P A. J Bacteriol. 1997;179:2740–2747. doi: 10.1128/jb.179.8.2740-2747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tauxe R, Blake P, Olsvik O, Wachsmuth K. In: The Future of Cholera: Persistence, Change, and An Expanding Research Agenda. Wachsmuth K, Blake P A, Olsvik O, editors. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 443–453. [Google Scholar]
- 47.Siddique A K, Zaman K, Akram K, Mutsuddy P, Eusof A, Sack R B. Trop Geogr Med. 1994;46:147–150. [PubMed] [Google Scholar]
- 48.Colwell R R. Science. 1996;274:2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- 49.Kolter R, Losick R. Science. 1998;280:226–227. doi: 10.1126/science.280.5361.226. [DOI] [PubMed] [Google Scholar]
- 50.O’Toole G A, Kolter R. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 51.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 52.Potera C. Science. 1996;273:1795–1797. doi: 10.1126/science.273.5283.1795. [DOI] [PubMed] [Google Scholar]