Abstract
The olfactory system is remarkable in its capacity to discriminate a wide range of odorants through a series of transduction events initiated in olfactory receptor neurons. Each olfactory neuron is expected to express only a single odorant receptor gene that belongs to the G protein coupled receptor family. The ligand–receptor interaction, however, has not been clearly characterized. This study demonstrates the functional identification of olfactory receptor(s) for specific odorant(s) from single olfactory neurons by a combination of Ca2+-imaging and reverse transcription–coupled PCR analysis. First, a candidate odorant receptor was cloned from a single tissue-printed olfactory neuron that displayed odorant-induced Ca2+ increase. Next, recombinant adenovirus-mediated expression of the isolated receptor gene was established in the olfactory epithelium by using green fluorescent protein as a marker. The infected neurons elicited external Ca2+ entry when exposed to the odorant that originally was used to identify the receptor gene. Experiments performed to determine ligand specificity revealed that the odorant receptor recognized specific structural motifs within odorant molecules. The odorant receptor-mediated signal transduction appears to be reconstituted by this two-step approach: the receptor screening for given odorant(s) from single neurons and the functional expression of the receptor via recombinant adenovirus. The present approach should enable us to examine not only ligand specificity of an odorant receptor but also receptor specificity and diversity for a particular odorant of interest.
The olfactory systems of vertebrates have a remarkable capacity to recognize and discriminate thousands of different odorant molecules (1–5). This process of discrimination, which results in sensory perception, begins with a series of signal transduction steps that occur within the olfactory neurons. The initial step in the process of odorant perception is the recognition of volatile odorant molecules by odorant receptors, members of the G protein coupled receptor superfamily, that are expressed on the surface of olfactory neuronal cilia (6, 7). The ligand(s)–receptor interactions then cause an increase in concentration of the intracellular messenger, cAMP, or inositol trisphosphate (1–5). Odorant-evoked elevations in cAMP are thought to directly activate a cation-selective cyclic nucleotide-gated channel, which causes external Ca2+ influx, leading to membrane depolarization and the generation of action potentials.
It is widely accepted that each olfactory neuron selectively expresses only one of hundreds of odorant receptor genes. Olfactory neuron cells expressing a given receptor are randomly dispersed within one of four distinct zones found in the olfactory epithelium (8, 9). The receptive range of each olfactory neuron is thought to be determined by the specific odorant receptor expressed by the neuron (10). Convergence of olfactory neurons expressing the same receptor onto two specific glomeruli (11, 12) suggests that the function of the glomerulus, which is characterized by the type of olfactory neurons connected to it, is to achieve finer tuning for given odorants at the level of the olfactory bulb (4). This notion is consistent with physiological studies of recordings taken from mitral cells responding to stimulation by various odorants (13). The link between the unique expression patterns of odorant receptors and the precise topographic map of olfactory neurons likely allows the olfactory system to establish the complex tuning mechanisms necessary for distinguishing a wide variety of structurally diverse odorants.
How is the enormous diversity of aromas recognized by only hundreds of odorant receptors? To understand the nature of odorant–odorant receptor interactions, several attempts have been made to determine the ligand specificity of individual receptor types. The odorant sensitivity was determined for a rat odorant receptor, OR5, which was expressed in Sf9 cells (7) or Escherichia coli (14), and a nonmammalian odorant receptor, the nematode ODR10, which was expressed in mammalian tissue cultures (15). Most recently, a rat odorant receptor I7 was functionally expressed by using an adenovirus-mediated expression system (16). Although these functional expressions of cloned odorant receptors allowed the determination of the chemical receptive ranges of receptor molecules in olfactory neurons, the conventional ligand screening steps have been a laborious task because of the enormous range of odorants. To overcome this problem, we herein demonstrate the functional cloning of an odorant receptor gene in an agonist-directed manner from single olfactory neurons by combining Ca2+ recording and single cell reverse transcription–coupled PCR techniques. Next, we adopt an adenovirus-mediated expression system to reconstitute the cloned receptor to ascertain the reliability of this approach and to enable further biochemical analyses. The present studies have tremendous potential to examine specificity and diversity of odorant receptors that recognize a particular odorant of interest.
MATERIALS AND METHODS
Isolation of Olfactory Receptor Neurons.
Olfactory epithelium was dissected out into Ringer’s solutions (138 mM NaCl/5.6 mM KCl/2 mM CaCl2/2 mM MgCl2/2 mM sodium pyruvate/9.4 mM glucose/5 mM Hepes, pH 7.4) from 3- to 4-week-old BALB/c CrSlc mice (Japan SLC, Hamamatsu, Japan) that were anesthetized by mioblock injection containing 20 μg/ml of bromopancronium (Sankyo). The epithelium pieces were treated with Ca2+-free Ringer’s for 10 min, 0.025% trypsin (Sigma) for 9–11 min at 37°C, 0.025% trypsin inhibitor (Sigma) for 10 min, and 0.1 mg/ml DNase (Sigma) for 2.5 min. The trypsinized olfactory epithelium cells then were tissue-printed on Cell-TAK (Collaborative Biomedical Products, Bedford, MA)-coated cover glass as described (10). In brief, superficial quasidissociated cells of the treated pieces were printed continuously on the coated glass floor by rolling the piece so that the lateral side maintained gentle contact with the floor by the use of an elastic glass pipette.
Fura-2 Ca2+-Imaging of Odorant Responses.
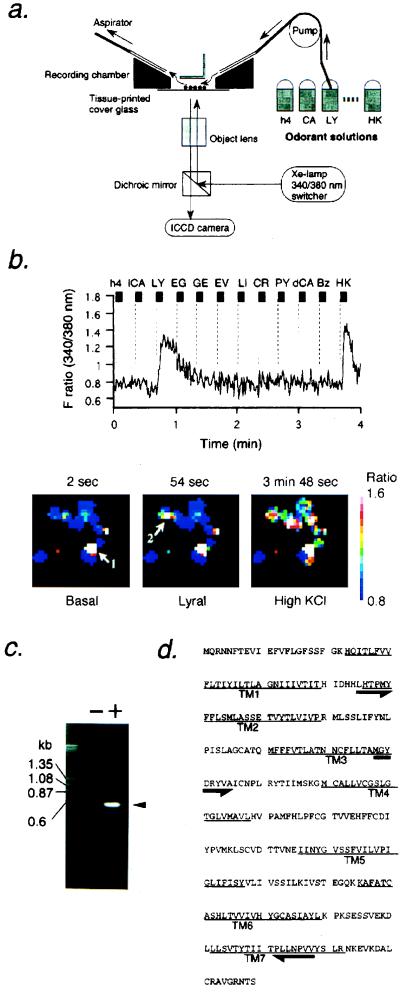
The cover glass was mounted on a teflon recording chamber, and 5 μM fura-2/AM (Molecular Probes) was loaded in tissue printed cells for 30 min. The fura-2 fluorescence at 510 nm by excitation at 340 or 380 nm was measured by use of inverted fluorescent microscope [Nikon TMD300 or Olympus (New Hyde Park, NY) IMT-2] and ICCD camera (Hamamatsu Photonics, Hamamatsu, Japan). Samples were illuminated by a xenon lamp, and a computer-controlled filter changer was used to switch the excitation wavelength. Images were recorded at 1.3-sec intervals and were analyzed with argus50 Ca2+-imaging processing system (Hamamatsu Photonics). Odorant solutions were applied to cells for 5–10 sec at indicated interstimulus intervals to wash the chamber solution. The flow rate was 1.5 ml/min with 50 μl of perfused volume of the chamber. This fura-2 Ca2+-imaging set-up is shown in Fig. 1a.
Figure 1.
Isolation of olfactory receptor gene from tissue printed olfactory neurons that respond to odorants using Ca2+ imaging and single cell RT-PCR. (a) Schematic drawing of Ca2+ imaging set up to record odorant-induced Ca2+ increase in tissue-printed olfactory neurons shown by circle dots. Odorant solutions were applied sequentially to the recording chamber with a peristaltic pump at the flow rate of 1.5 ml/min. (b) The response of a single olfactory neuron to lyral (shown by arrow 2 in lower panel) in reflected changes in fura-2 fluorescence intensity ratios (340/380 nm). The odorants were applied to tissue-printed cells for 4 sec during times indicated by the bars. The cells were washed continuously between odorant applications. Names of odorants are abbreviated as follows: h4, 1-butanol; lCA, l-carvone; LY, lyral; EG, eugenol; GE, geraniol; EV, ethyl vanillin; LI, lilial; CR, cresol; PY, pyridine; dCA, d-carvone; Bz, benzene (each 100 μM). HK stands for high KCl buffer (145.6 mM KCl/2 mM CaCl2/2 mM MgCl2/9.4 mM glucose/5 mM Hepes, pH 7.4). Lower panels show pseudocolored images of Ca2+ measurements taken from tissue-printed cells at three representative time-points. The Ca2+ level of the lyral-responding cell (arrow 2) was increased 17-fold on lyral application from 246 nM (a basal level) to 4.2 μM. Arrow 1 depicts a dead cell. The graded color bar is a calibration of the imaging system. The white color indicates the highest Ca2+ level while the blue color represents the basal level and outlines the shape of each cell. (c) Single cell RT-PCR products separated on agarose gel. Minus and plus signs indicate RT-PCR experiments with or without reverse transcriptase using the same RNA preparations. (d) The putative amino acid sequence of the MOR23 coding region. The primers used for two-round PCR are shown by arrows. TM, putative transmembrane domains.
Single Cell RT-PCR.
The odorant-responsive single cell was isolated in a glass capillary tube (GC150–10, Clark Electromedical Instruments, Pangbourne, U.K.), which then was ejected into guanidine isothiocyanate solution. The crude RNA was purified through GlassMAX spin cartridge (GIBCO/BRL) in the presence of tRNA as a carrier. The RNA was eluted with preheated RNase free water, which then was treated with DNase to remove genomic DNA. After DNase was inactivated, the sample was divided into two; one aliquot was subjected to reverse transcriptase reaction in 100 units of RNaseH− Superscript II (GIBCO/BRL), 25 mM Tris⋅HCl, 50 mM KCl, 2.5 mM MgCl2, 6.7 mM DTT, 12 units of RNasin (Promega), 82 nM d(T)18-NotI (Pharmacia), and 0.34 mM dNTP, and another was without the Superscript II enzyme as a control. The mixture was incubated for 50 min at 42°C, and the reaction was stopped by heating for 15 min at 65°C. Quality of the cDNA was verified by amplification of a cDNA fragment of glucose-3-phosphate dehydrogenase that was ubiquitously expressed in essentially all tissues. The first amplification of olfactory receptor gene was performed in a solution containing 0.4 μM each of a degenerate primer derived from olfactory receptor gene (5′ (T/C)TIC(T/C)ACICCIATGTA 3′, corresponding to the primer in TM2 shown in Fig. 1d) and a NotI-adapter primer designed from d(T)18-NotI that was used for the first strand cDNA synthesis, 0.2 mM dNTP, and AmpliTaq Gold (2.5 units/50 μl) according to the following schedule: the first 10-min treatment at 95°C followed by 40 PCR cycles at 95°C for 1 min, various annealing temperatures depending on primers for 3 min, and 72°C for 2 min by using RoboCycler (Stratagene) or TaKaRa PCR thermal cycler MP (TaKaRa Biomedical, Shiga, Japan) with sample temperature monitoring. The second amplification was done with the same schedule by use of a set of primers indicated in Fig. 1d [5′ ATGGCITA(C/T)GA(C/T)(A/C)GITA(C/T)GTIGC 3′ (TM3) and 5′ AT(G/A)AAIGG(G/A)TTIA(G/A)CATIGG 3′ (TM7)]. The PCR products were subsequently TA-cloned into pCR (Invitrogen), pGEM-T (Promega), or pT7Blue-T (Novagen) vectors, followed by sequencing analysis.
Molecular Cloning of the Mouse Olfactory Receptor (MOR) 23 Coding Sequence.
The entire coding region of MOR23 was amplified by PCR from the mouse MOR23 genomic clone with the upstream primer 5′-CAGACCGTGCATCATGCAGAGAAATAACTTC-3′ and the downstream primer 5′-GATATCAAGAAGTGTTTCTGCCCA-3′. The amplified fragment then was cloned into pGEM-T vector (Promega), and the MOR23 sequence was verified by sequencing.
Construction of Expression Units.
Before construction, the usefulness of both the adenovirus vector and green fluorescent protein (GFP) to the olfactory neurons was evaluated by use of a GFP-expressing adenovirus vector (17). To generate the monocistronic expression unit for GFP, the coding sequence for an enhanced GFP derived from pEGFP-N1 (CLONTECH) was subcloned into a cosmid vector pAxCAwt (18). To generate a bicistronic expression unit for MOR23 and GFP, the MOR23 coding region followed by the internal ribosome entry site (IRES) derived from pCITE-1 (Novagen) (19) and the GFP coding region were subcloned into a cosmid vector pAxLNLw (18). For each were construct, the expression unit contains CAG promoter (20), and the cosmid carrying the insert in the proper orientation was selected.
Generation and Application of the Adenovirus Vectors.
Recombinant adenovirus vectors were generated by cosmidterminal protein complex method as described (21). Recombinant adenovirus vectors then were amplified further to larger scale in HEK293. For the bicistronic adenovirus vector, the expression of the bicistronic unit was turned on by a Cre-expressing adenovirus vector AxCANCre (18) via a site-directed recombination at the final step of amplification, which was confirmed by Southern blot analysis. A total of 2.5–5.0 μl of adenovirus vector solution at a titer of 109 pfu/ml was injected by pipetting at 0.25–0.5 μl/min into a nostril of 3–4-week-old C57BL/6NCrj mice (Charles River Breeding Laboratories).
Tissue Observation.
Infected or uninfected C57BL/6NCrj mice were killed under perfusion of 4%-paraformaldehyde in PBS (pH7.4). Dissected tissues were fixed with 4% paraformaldehyde in PBS (pH7.4) at 4°C for 60 min to 1 day, were decalcinated with 500 mM EDTA for 1–2 day(s) at 4°C, and were immerged with 30% sucrose for 4–16 hours at 4°C. The pretreated tissues were overspread in OCT compound (Miles Scientific), were frozen in dry ice–acetone, and then were cut into 10–20 μm cryosections. In situ hybridization was performed as described elsewhere (22). Labeled antisense RNA probes were prepared with digoxigenin-11-UTP by using DIG DNA Labeling Kit (Boehringer Mannheim) according to the manufacturer’s instructions. For observation of GFP fluorescence, sections were dipped in a solution containing 10 mM Tris⋅HCl and 1 mM EDTA (pH8.0) and were observed under fluorescent microscopy.
RESULTS AND DISCUSSION
Discrimination of molecular signals in the olfactory system consists of the initial reception of odorants by olfactory neurons followed by the subsequent processing in the olfactory bulb and olfactory cortex. Molecular receptive range in olfactory neurons and bulbs are based on the information coded within odorant molecules such as differences in chain length, terminal groups, and positions of functional groups (4, 10). Direct insight into odorant–olfactory receptor interactions, however, has not yet been obtained because of the difficulty in expressing functional receptors necessary for the screening of thousands of odorant molecules. To approach a detailed understanding of the dynamic tuning mechanisms involved in discriminating a variety of odorants at receptor level, we reasoned that the combination of odorant response assay with single cell RT-PCR analysis (23) would lead to the determination of the types of receptors expressed by single olfactory neurons and a correlation with the physiological responses to those receptors.
Olfactory receptor neurons suitable for simultaneous recording of odorant responses in several cells were isolated by use of tissue printing method (10) from mouse olfactory epithelium located on the upper septum and roof designated as zone 1 among four distinct spatial receptor zones (9). Odorant receptor-mediated elevations in cAMP or inositol-1,4,5-trisphosphate trigger Ca2+ influx through a cation-selective cyclic nucleotide-gated channel or an as-yet uncharacterized inositol trisphosphate-gated ion channel, respectively (1–5). Tissue-printed olfactory neurons were thus subjected to fura-2 based Ca2+imaging to measure increases in intracellular Ca2+ levels in response to odorant stimulations. Sequential applications of various odorants by use of the recording chamber shown in Fig. 1a allowed identification of cells responding to certain odorant(s).
For example, shown in Fig. 1b is a Ca2+-level recording of a single cell that responds only to the odorous aldehyde, lyral, at 100 μM concentrations among 11 sequentially applied odorants (the structure of lyral is shown in Fig. 4). Viability of the cell was confirmed by high potassium chloride buffer-induced membrane depolarization. Of ≈3,500 tissue-printed viable olfactory neurons, we could identify a total of 226 positive cells that responded to some of the 11 applied odorants. For example, 29 cells responded to eugenol, 44 responded to ethylvanillin, 2 responded to geraniol, 16 responded to lilial, 41 responded to cresol, and 6 responded to lyral. Some cells showed responses to more than one odorant (data not shown). Each responsive cell was subsequently picked in a microcapillary tube and was subjected to single cell RT-PCR using degenerate oligonucleotides designed on the basis of the known conserved sequences among the olfactory receptor superfamily (6, 7). The DNase treatment of column-purified RNA samples and the first PCR by using the oligo(dT)-adapter sequence eliminated contaminating PCR products derived from genomic DNA (Fig. 1c). Thus, no PCR product was observed in the sample that had not undergone the reverse transcriptase reaction (Fig. 1c). Sequence analysis of the two-round PCR-amplified fragment obtained from one lyral responsive cell (Fig. 1b) revealed it to be identical to the mouse olfactory receptor MOR23 (24) (Fig. 1d). Consistent with previous implication (8, 9), it was found that only one type of odorant receptor was isolated from a single cell. We also have isolated 15 additional odorant receptor genes from cells that responded to other odorants used in this study such as eugenol and cresol, although the characterization of these single cell RT-PCR products is in progress.
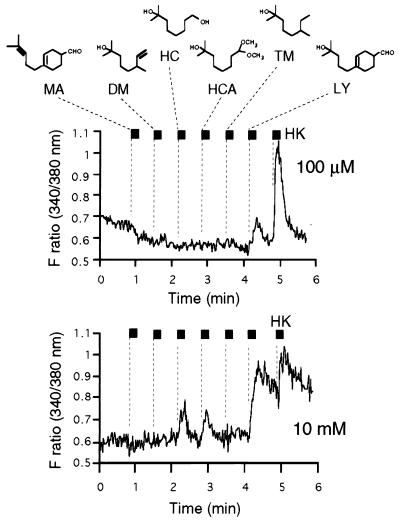
Figure 4.
Ligand specificity of MOR23. Ca2+ increase in MOR23-expressing cells was induced by some odorants structurally similar to lyral at higher concentrations. MA, myrac aldehyde; DM, dihydromyrcerol; HC, hydroxycitronellol; HCA, hydroxycitronellol dimethyl acetal; TM, tetrahydromyrcenol. MOR23 responds to HC and HC dimethyl acetal (HCA) at 10 mM but not at 100 μM. Common structural motifs are shown by boxes. LY is a mixture of two isomers: 4-(4-hydroxy-4-methylpentyl)-3-cyclohexenecarbaldehyde and 3-(4-hydroxy-4-methylpentyl)-3-cyclohexenecarbaldehyde. MA is also a mixture of two isomers, which are the dehydrated forms of LY (hydroxy MA). Only 4-substituted isomers of LY and MA are depicted in the figure.
Among them, the MOR23 is of particular interest because it has been relatively well characterized and has distinct regulatory expression patterns in olfactory neurons and testis tissues (24, 25). It was confirmed by an in situ hybridization experiment that the expression of the endogenous MOR23 is restricted to epithelium of the upper septum and roof defined as expression zone 1 (9), the same region used for the tissue printing (Fig. 2a). At this stage, although lyral was a candidate ligand for MOR23, a functional reconstitution study was required to verify lyral-MOR23 interaction.
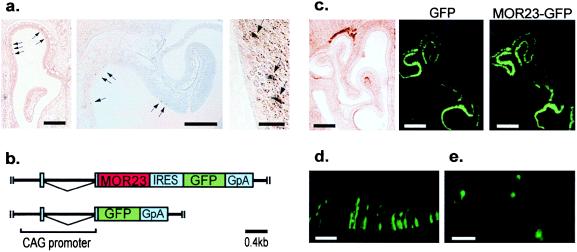
Figure 2.
Localization of the endogenous MOR23 gene and expression of the recombinant adenovirus-mediated transgene in olfactory epithelium. (a) In situ hybridization of the MOR23 transcript in coronal sections of the olfactory epithelium. The antisense RNA for the MOR23 coding region was used as a probe. The presence of the transcripts was indicated by dispersed focal staining shown by arrows. The expression of MOR23 is totally restricted to the expression zone 1 at every section. [Bars = 0.5 mm (left), 250 μm (middle), and 100 μm (right).] (b) Schematic drawings of the expression units of the recombinant adenovirus vectors used in this study. Regions corresponding to the mature transcripts are indicated by boxes. The bicistronic adenovirus expression unit (Upper) contains MOR23 gene, IRES sequence, and GFP gene followed by the rabbit β-globin polyadenylation signal (GpA). The monocistronic expression unit (Lower) contains the GFP gene followed by GpA. The IRES allows internal translation initiation, resulting in expression of both MOR23 and GFP. These individual expression units are located at the E1A-deleted region of the adenovirus type-5 genome. (Bar = 0.4 kilobases). (c) Coronal sections of the nasal tissues infected with the monocistronic (Left and Center) and the bicistronic (Right) recombinant adenovirus vectors. (Left) Under light microscopy. (Center and Right) Under fluorescent illumination. There are no significant differences in the infected regions throughout the olfactory epithelium between two constructs. (Bars = 0.5 mm.) (d) Higher magnification of MOR23 virus-infected cells. The fluorescent cells are predicted to be olfactory neurons. (Bar = 100 μm.) (e) Tissue-printing of the GFP fluorescent olfactory epithelium result in identification of MOR23-infected individual single olfactory neurons under fluorescence illumination. (Bar = 100 μm.)
We then targeted the olfactory neuron itself as an expression system for the reconstitution of the odorant-receptor interaction. Thus, recombinant adenovirus vectors containing a bicistronic expression unit for MOR23 and GFP, and a monocistronic one for GFP alone as a negative control vector, were constructed (Fig. 2b). When these vectors were applied to nasal tissues, the presence of the GFP protein was detectable in both cases by fluorescent microscopy (Fig. 2 c and d). In each infection—i.e., the bicistronic MOR23 vector and the monocistronic GFP vector—coronal sections in the middle part of the epithelium exhibited robust fluorescence within the dorsolateral recessed area at the confronted surfaces between the turbinates and lower septum, none of which is a part of zone 1, where the endogenous MOR23 is expressed. The same observations were obtained throughout the anterior–posterior axis (data not shown), thereby ruling out the possibility for a meeting of the endogenous MOR23 and the transgene in the same cell. There was no significant difference in expressed zones between the MOR23 and GFP constructs (Fig. 2c), suggesting that the extent of the viral infection seems to depend on how much longer the peripheral space can retain the viral solution within its cavity.
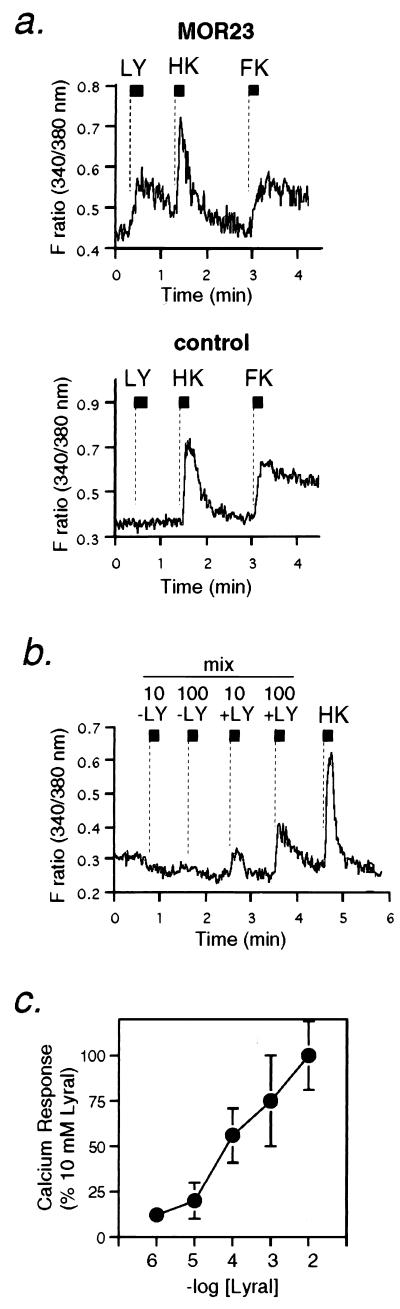
To test odorant responses at single cell level, the MOR23 adenovirus-infected fluorescent cells were individually isolated by the tissue printing method from the GFP-fluorescent epithelium within zone 2 that lay immediately ventral or lateral to zone 1 in which the endogenous MOR23 gene is expressed (Fig. 2e). In Ca2+-level recordings of single infected neurons, we could detect responses to lyral (1 mM) in 16 of 22 GFP-positive cells that were also sensitive to both high KCl buffer and forskolin, a reagent that increases cyclic AMP levels (Fig. 3a Upper). In contrast, no response to lyral was observed in the monocistronic GFP-infected cells similarly isolated from the same zone in control mice (n = 17) (Fig. 3a Lower). The 40–50% of MOR23 adenovirus-infected cells that did not respond to forskolin also did not elicit lyral-responses (n = 19) (Table 1), suggesting that the failure of responding to lyral and forskolin in these cells resulted from a common cause.
Figure 3.
Characterization of odorant responses in MOR23-adenovirus infected cells. (a) Lyral-mediated Ca2+ increase in MOR23 adenovirus-infected cells shown by changes in fura-2 fluorescence intensity ratios (340/380 nm) (1 mM lyral application for 10 sec at the indicated bars). (Upper) Bicistronic MOR23-IRES-GFP. (Lower) Monocistronic GFP. High KCl buffer (HK) and forskolin (FK) (10−5 M) were applied at indicated bars (5 sec) to assess cell viability and olfactory neuronal properties. (b) Ca2+ increase in MOR23-adenovirus infected cells was induced by lyral-containing odorant mixture (mix + LY) but not by the mixture without the lyral (mix − LY). The mixture contains the same odorants used in Fig. 1b (10 or 100 μM). (c) Dose-dependent Ca2+ increase as a percentage of the response at 10 mM lyral. Data ±SE (n = 16 for 1 mM, n = 5 for others).
Table 1.
Number of infected cells responding to lyral
| high KCl* | Forskolin† | Lyral response | |
|---|---|---|---|
| GFP | + | + | 0 (n = 17) |
| + | − | 0 (n = 14) | |
| MOR23 | + | + | 16 (n = 22) |
| + | − | 0 (n = 19) | |
| +Thapsigargin | + | + | 7 (n = 11) |
GFP, monocistronic GFP adenovirus vector; MOR23, bicistronic MOR23-IRES–GFP vector.
High K+-responsive viable cells are 30–60% of total tissue-printed fluorescent cells, depending on preparation.
The 50–60% of high K+-responsive cells elicit Ca2+ increase on forskolin stimulation (10−5 M).
To assess whether the ligand specificity of reconstituted MOR23 resembles that of the endogenous MOR23, the odorant mixture containing the same compounds used in Fig. 1b (10 or 100 μM) was applied to the infected cells. The MOR23 adenovirus-infected cells responded only to those odorant cocktails containing lyral in a dose-dependent manner (Fig. 3b). The dose-dependent responses to lyral are observed at concentrations of 10−5–10−2 M (Fig. 3c). These reconstitution studies indicate that the MOR23 receptor indeed interacts with the lyral molecule and transmits signals leading to the intracellular Ca2+ increase. Attempts were made to identify other ligands for MOR23 based on two functional units in lyral molecules, i.e., the tertiary alcohol and aldehyde groups (Fig. 4). Myrac aldehyde (MA) contains cyclohexenecarbaldehyde moiety as a common motif with lyral. Dihydromyrcerol, hydroxycitronellol (HC), HC dimethyl acetal, and tetrahydromyrcenol share a common tertiary alcohol unit in lyral molecules. None of these compounds was recognized by MOR23 in Ca2+-responsive assays at either 100 μM (Fig. 4) or 1 mM concentrations (data now shown) except lyral whereas HC and HC dimethyl acetal evoked Ca2+ increase at 10 mM concentrations (Fig. 4). These results implicate that ligand specificity of MOR23 is highly defined to the presence of both functional units in lyral, although the main functional group recognized by the MOR23 binding pocket is the tertiary alcohol moiety. It should be noted, however, that quantitative comparison of the affinities of these ligands is difficult because of the differences in solubilities of these compounds.
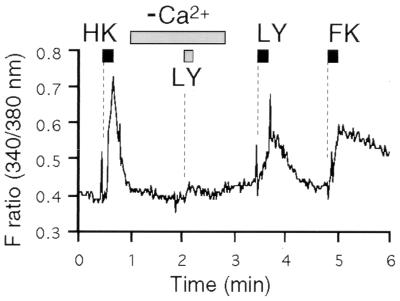
To determine the origin of Ca2+ increase triggered by lyral-MOR23 interaction, Ca2+ recordings in infected neurons were performed in the absence of external Ca2+. Forskolin-mediated Ca2+ increase was significantly impaired without external Ca2+ (data not shown). Similarly, the Ca2+-free condition completely abolished the lyral-MOR23-mediated elevation of intracellular Ca2+ whereas the same cell responded to lyral after the exchange of buffer to Ca2+ containing Ringer’s solution (Fig. 5). These results indicate that lyral binding to the MOR23 receptor leads to external Ca2+ influx through cell surface channels. The treatment of the cells with thapsigargin, a Ca2+-ATPase inhibitor that depletes intracellular Ca2+ stores, did not affect the Ca2+ increases induced by lyral, high KCl-buffer, or forskolin, further confirming that lyral-MOR23-mediated Ca2+ increase is not derived from internal Ca2+ stores (Table 1).
Figure 5.
Lyral-MOR23 interaction leads to external Ca2+ influx. Lyral-induced Ca2+ increase is greatly reduced in the absence of external Ca2+. The buffer was exchanged to Ca2+-free Ringer’s solution for the indicated period, during which lyral in Ca2+-free buffer was applied. The response was rescued by changing the buffer to Ca2+-containing Ringer’s solution.
Conventional expression studies to screen ligands recognized by a receptor of interest have been a laborious task in the case of olfactory receptors because of the vast range of odorant molecules. In this study, functional expression and ligand screening of odorant receptor was greatly facilitated by the fact that the candidate ligand was already specified during the functional identification of the receptor from single olfactory neurons. In addition, our approach enables the screening of receptors for an odorant of interest at single cell level. Therefore, isolation of several receptors, which respond to a certain odorant, and their homology search will provide information on not only ligand-specificity of the receptors but also aspects of evolution of thousands of olfactory receptor proteins. We have not been able to identify additional odorant receptor genes from cells responding to lyral. But it is reasonable to think that there exist more receptors that recognize lyral with different ligand specificities. Further analysis of lyral responsive cells is necessary to address structural aspects of the ligand-receptor interaction and tuning mechanisms that allow determination of the specific aroma.
In addition to the MOR23 gene, we have so far cloned 15 other odorant receptor genes from the 220 cells that responded to some of the 11 tested odorants such as eugenol, cresol, and ethyl vanillin. Reconstitutions of these receptors are currently in progress. There are a couple of reasons for being unable to isolate odorant receptors from all responsive cells. First, we had to eliminate ≈25% of samples because of contamination by neighboring cells during picking up of the responsive cell. Second, according to the PCR analyses using glucose-3-phosphate dehydrogenase, RNA isolation and subsequent reverse transcription were not successfully performed in ≈40% of samples. Third, even though we used degenerate primers designed on the basis of known odorant receptor genes, it is highly possible that not all odorant receptor genes can be amplified with the primers used in this study. The use of other primer sets could increase the efficiency of single cell RT-PCR.
The adenovirus-mediated expression system was used similarly for a rat odorant receptor I7 by Zhao et al. (16). They used the system to identify ligands to the orphan receptor whereas the purpose of our study in using the adenovirus approach was to reconstitute the odorant response observed in the original olfactory neuron and to confirm the correlation of the cloned receptor with the odorant response. Both their study and ours clearly support the notion that an adenovirus vector is a useful tool to functionally express odorant receptors in the olfactory neurons. Caution has to be taken herein to avoid the presence of the endogenous receptor for the odorant under study. In our study, the number of the neurons expressing the endogenous MOR23 gene is relatively small, and the regions expressing the transgene were clearly distinct from those of the endogenous receptor. Thus, the possibility of mistaking endogenous signaling in infected cells was minimized.
In summary, this study demonstrates the functional identification of odorant receptor from single olfactory neurons, using Ca2+ imaging and single cell RT-PCR followed by functional expression of the receptor using bicistronic recombinant adenoviruses. The reconstitution of MOR23 in olfactory receptor neurons proved that the lyral response, which was originally observed in the tissue-printed single olfactory neuron, was indeed derived from endogenous MOR23-lyral interaction and that the type of an odorant receptor expressed by a single olfactory neuron correlates with the physiological response of the cell to the specific odorants. MOR23 also recognized compounds structurally related to lyral, suggesting that odorant receptors possess a receptive range based on structural determinants in odorant molecules. This strategy for identifying receptors for a particular odorant of interest should prove to be a powerful method for studying complex odorant(s)–receptor interactions. The present approach also could apply to other receptor systems in which potential subtypes of the receptor are differentially expressed in tissues containing heterogeneous cell types. Further, agonist-directed screening of the receptor could lead to identification of new receptor subtypes with different pharmacological properties. Currently, we are screening more receptors from single neurons that respond to specific odorant(s) to define tuning mechanisms that allow various odorants to be discriminated by the odorant receptors in the olfactory neurons.
Acknowledgments
We thank Drs. T. Takahashi, T. Kurahashi, K. Mori, K. Tsuzuki, S. Ozawa, K. Abe, M. Yamagata, M. Noda, Y. Hamada, and M. Hashimoto for helpful suggestions. We thank Drs. I. Saito and T. Nakatsu for supplying materials for virus construction, Dr. J. Miyazaki for providing a CAG promoter, and Dr. K. Moriyoshi for an GFP adenovirus vector. Special thanks are due to Dr. F. Nagawa and other lab members for discussion. We also thank Dr. D. W. Saffen and Ms. J. Geng for English corrections. We thank the T. Hasegawa Co., Ltd., for odorant compounds. This work was supported by Grant-in-Aid for Scientific Research-C and on Priority Areas (to K.T.), for Specially Promoted Research (to H.S.), and for Research for the Future of Japan Society for the Promotion of Science (to T.H.) from the Ministry of Education, Science, Sports, and Culture, by CREST of Japan Science and Technology Corporation (to T.H.), and by grants from the Ministry of International Trade and Industry (to T.S. and K.T.).
ABBREVIATIONS
- RT-PCR
reverse transcription–coupled PCR
- GFP
green fluorescent protein
- MOR
mouse olfactory receptor
- MA
myrac aldehyde
- HC
hydroxycitronellol
- IRES
internal ribosome entry site
Note Added in Proof
After this paper was submitted, Krautwurst el al. (26) reported functional expression of a chimeric–receptor library in HEK293. Both their approach and ours enable us to study receptor–odorant interactions by providing a system for isolating receptors that are specific to a certain odorant molecule.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Buck L B. Annu Rev Neurosci. 1996;19:517–544. doi: 10.1146/annurev.ne.19.030196.002505. [DOI] [PubMed] [Google Scholar]
- 2.Shepherd G M. Neuron. 1994;13:771–790. doi: 10.1016/0896-6273(94)90245-3. [DOI] [PubMed] [Google Scholar]
- 3.Reed R R. Neuron. 1992;8:205–209. doi: 10.1016/0896-6273(92)90287-n. [DOI] [PubMed] [Google Scholar]
- 4.Mori K, Yoshihara Y. Prog Neurobiol. 1995;45:585–620. doi: 10.1016/0301-0082(94)00058-p. [DOI] [PubMed] [Google Scholar]
- 5.Lancet D, Ben-Arie N. Curr Biol. 1993;3:668–674. doi: 10.1016/0960-9822(93)90064-u. [DOI] [PubMed] [Google Scholar]
- 6.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 7.Raming K, Krieger J, Strotmann J, Boekhoff I, Kubick S, Baumstark C, Breer H. Nature (London) 1993;361:353–356. doi: 10.1038/361353a0. [DOI] [PubMed] [Google Scholar]
- 8.Ressler K J, Sullivan S L, Buck L B. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 9.Vassar R, Ngai J, Axel R. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- 10.Sato T, Hirono J, Tonoike M, Takebayashi M. J Neurophysiol. 1994;72:2980–2989. doi: 10.1152/jn.1994.72.6.2980. [DOI] [PubMed] [Google Scholar]
- 11.Vassar R, Chao S K, Sitcheran R, Nunez J M, Vosshall L B, Axel R. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 12.Ressler K J, Sullivan S L, Buck L B. Cell. 1994;79:1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 13.Mori K. J Neurophysiol. 1992;67:786–789. doi: 10.1152/jn.1992.67.3.786. [DOI] [PubMed] [Google Scholar]
- 14.Kiefer H, Krieger J, Olszewski J D, von Heijne G, Prestwich G D, Breer H. Biochemisty. 1996;35:16077–16084. doi: 10.1021/bi9612069. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Chou J H, Bradley J, Bargmann C I, Zinn K. Proc Natl Acad Sci USA. 1997;94:12162–12167. doi: 10.1073/pnas.94.22.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao H, Lidija I, Otaki J M, Hashimoto M, Mikoshiba K, Firestein S. Science. 1998;279:237–242. doi: 10.1126/science.279.5348.237. [DOI] [PubMed] [Google Scholar]
- 17.Moriyoshi K, Richards L J, Akazawa C, O’Leary D D, Nakanishi S. Neuron. 1996;16:255–260. doi: 10.1016/s0896-6273(00)80044-6. [DOI] [PubMed] [Google Scholar]
- 18.Kanegae Y, Lee G, Sata Y, Tanaka M, Nakai M, Sakaki T, Sugano S, Saito I. Nucleic Acids Res. 1995;23:3816–3821. doi: 10.1093/nar/23.19.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D G, Kang H M, Jang S K, Shin H S. Mol Biol Chem. 1992;12:3636–3643. doi: 10.1128/mcb.12.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 21.Miyake S, Makimura M, Kanegae Y, Harada S, Sato Y, Takamori K, Tokuda C, Saito I. Proc Natl Acad Sci USA. 1996;93:1320–1324. doi: 10.1073/pnas.93.3.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirota S, Ito A, Morii E, Wanaka A, Tohyama M, Kitamura Y, Nomura S. Mol Brain Res. 1992;15:47–54. doi: 10.1016/0169-328x(92)90150-a. [DOI] [PubMed] [Google Scholar]
- 23.Sucher N J, Deitcher D L. Neuron. 1995;14:1095–1100. doi: 10.1016/0896-6273(95)90257-0. [DOI] [PubMed] [Google Scholar]
- 24.Asai H, Kasai H, Matsuda Y, Yamazaki N, Nagawa F, Sakano H, Tsuboi A. Biochem Biophys Res Commun. 1996;221:240–247. doi: 10.1006/bbrc.1996.0580. [DOI] [PubMed] [Google Scholar]
- 25.Parmentier M, Libert F, Schurmans S, Schiffmann S, Lefort A, Eggerick D, Ledent C, Mollereau C, Gerard C, Perret J, et al. Nature (London) 1992;355:453–455. doi: 10.1038/355453a0. [DOI] [PubMed] [Google Scholar]
- 26.Krautwurst D, Yan K W, Reed R R. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]