Abstract
ParM is a prokaryotic actin homologue, which ensures even plasmid segregation before bacterial cell division. In vivo, ParM forms a labile filament bundle that is reminiscent of the more complex spindle formed by microtubules partitioning chromosomes in eukaryotic cells. However, little is known about the underlying structural mechanism of DNA segregation by ParM filaments and the accompanying dynamic instability. Our biochemical, TIRF microscopy and high-pressure SAX observations indicate that polymerization and disintegration of ParM filaments is driven by GTP rather than ATP and that ParM acts as a GTP-driven molecular switch similar to a G protein. Image analysis of electron micrographs reveals that the ParM filament is a left-handed helix, opposed to the right-handed actin polymer. Nevertheless, the intersubunit contacts are similar to those of actin. Our atomic model of the ParM-GMPPNP filament, which also fits well to X-ray fibre diffraction patterns from oriented gels, can explain why after nucleotide release, large conformational changes of the protomer lead to a breakage of intra- and interstrand interactions, and thus to the observed disintegration of the ParM filament after DNA segregation.
Keywords: GTPase, left-handed helix, molecular switch, ParM
Introduction
In Escherichia coli plasmid R1 (Møller-Jensen et al, 2000), single-copy stability is accomplished by only three components: a filament-forming protein ParM, a DNA-binding protein ParR and a centromere-like site in the DNA (parC) (Gerdes et al, 2000). ParR–parC binding to ParM is thought to induce ParM polymerization, the filaments formed pushing the plasmids to opposing bacterial poles (Møller-Jensen et al, 2002). Despite some significant differences in loop, helix and sheet arrangement within the four subdomains (Ia, Ib, IIa and IIb) (van den Ent et al, 2002), the crystal structure of ParM proved to be similar to g-actin, confirming the original sequence-based assignment of an ‘actin fold' (van den Ent et al, 2002). Two different crystal forms revealed that on loss of ADP, a large conformational change (≈25°) occurs between subdomains I and II that results in the opening of the interdomain cleft (van den Ent et al, 2002). In vivo, ParM filaments are only visible in about 40% of a bacterial population at any given time point, indicating that they are transient and dynamically instable (Møller-Jensen et al, 2002). As a response to molecular crowding (about 30% of a bacterium is filled with various proteins), ParM filaments are usually observed as randomly oriented bundles both in vivo (Møller-Jensen et al, 2002) and in vitro (Popp et al, 2007c). The structural basis leading to the observed dynamic instability of ParM filaments remains unknown.
In this paper, on the basis of our results, we show that ParM, despite being a member of the actin family containing the typical structural motif ‘actin fold', which characterizes it as an ATPase, acts as a GTP-driven molecular switch, resembling some behaviour of G proteins.
We have characterized the nucleotide-dependent dynamics of ParM filaments by a wide variety of methods, including TIRF microscopy and high-pressure SAX. Together with our structural analysis of ParM, combining X-ray crystallography of the monomer, EM reconstructions of the filament and high-resolution X-ray diffraction patterns from oriented ParM gels, a picture has been emerged to reveal how the biochemical and structural instability of ParM are related.
Results
Probing nucleotide-dependent instability
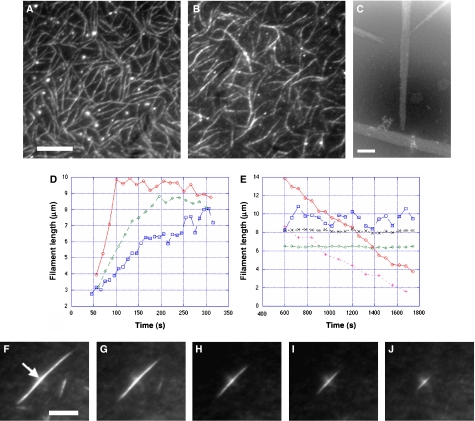
Using in vitro TIRF microscopy, we found that the addition of not only ATP (Garner et al, 2004; Popp et al, 2007c) but also GTP and its analogues leads to ParM filament formation (Figure 1A and B; Supplementary movie 1). Yet the dynamic properties of ParM filaments were highly dependent on the bound nucleotides. The polymerization rate was substantially faster with GTP than with ATP, whereas a (1:1) mixture of ATP and GTP (ParM-Mix filaments) resulted in a polymerization rate halfway in between (Figure 1D). At steady state, a more dramatic difference is seen. ParM filaments polymerized with nonhydrolysable nucleotides like ATPγS or GMPPNP were stable and retained a constant length. ParM-ATP filaments underwent phases of repeated catastrophe and rescue, similar to microtubules (Supplementary movie 2), whereas ParM-GTP and ParM-Mix filaments quite steadily disintegrated into monomers to completion (Figure 1E; Supplementary movies 3 and 4). Disintegration of subunits appeared to be approximately symmetric at either end of the bidirectional ParM bundles observed in our in vitro TIRF microscopy assays in the presence of crowding agents (Figure 1C, F–J; Supplementary movie 3).
Figure 1.
Typical ParM filaments observed in the TIRF microscope at steady state. (A) Polymerization of ParM (5.4 μM) was initiated by adding 10 mM ATP, in the presence of 8% PVA. (B) Same as (A) but 10 mM GTP was added to initiate polymerization (see Supplementary movie 1) Scale bar 5 μm. (C) In the electron microscope, the filaments observed under the TIRF microscope (A, B) usually appear as bundles in the presence of crowding agents. Conditions were 5.4 μM ParM, 10 mM GTP and 8% PVA. Scale bar, 100 nm. Note that ParM bundles look spindle-shaped for both ParM-GTP and ParM-ATP filaments, whereas filaments polymerized with nonhydrolysable nucleotides are straight with blunt ends (data not shown) (D) Typical time courses of polymerizing ParM filaments measured from TIRF microscopy images. Conditions were 5.4 μM ParM in the presence of 8% PVA. After adding 10 mM GTP (red), 10 mM ATP (blue) and a 1:1 mixture of GTP and ATP (10 mM Mix) in green symbols. (E) Typical behaviour of some filaments at steady state. ParM (4 μM) with 8% PVA. ParM-GTP filaments (red) and ParM-Mix filaments (pink) decreased steadily, ParM-ATP filaments (blue) displayed microtubule-like instability, ParM-ATPγS (black) and ParM-GMPPNP filaments (green) were stable filaments. (F–J) An example of a ParM-GTP bundle (5.4 μM, 8% PVA) disintegrating approximately symmetrically at both ends. Scale bar, 10 μm. At 240 s (F), 510 s (G), 810 s (H), 1110 s (I) and 1410 s (J) after initiating polymerization by adding 10 mM GTP.
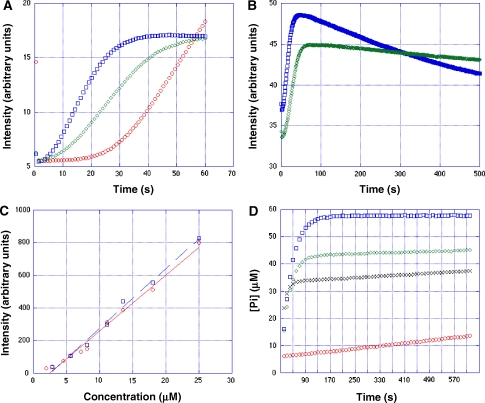
We confirmed the findings on the nucleotide-dependent polymerization and steady-state behaviour by stopped flow performed on crowding-agent-free buffers where ParM forms predominantly single filaments (Figure 2A and B; Supplementary Figure S9). The critical concentrations of ParM-ATP and ParM-GTP filaments were similar and both were in the range of 1.5–2 μM, as determined by both light scattering (Figure 2C) and TIRF microscopy (Supplementary Figure S11).
Figure 2.
(A) Monitoring the early phase of ParM (6 μM) polymerization by stopped flow at 25°C. ParM-GTP (blue), ParM-ATP (red) and ParM-Mix (green). The total amount of nucleotide was 5 μM. Intensities were scaled to the initial value of ParM-ATP. The unscaled raw experimental data over a long time range is shown in the Supplementary Figure S9. Note that the light-scattering intensity for both ParM-GTP (blue symbols) and ParM-Mix filaments (green symbols) indicated a biphasic behaviour as the light-scattering intensity jumped up to a value of about 35% in the first two frames, whereas for ParM-ATP (red symbols) the initial light-scattering intensity was only about 5%. This indicates that when adding GTP, there is a very rapid initial polymerization of ParM within the first second of mixing and too fast for us to resolve at the present stage, followed by a second phase, which is still very rapid compared with ATP. The scaled data highlights the second phase. (B) Same experiment as in (A) but now highlighting the decaying phase of ParM-GTP (blue symbols) and ParM-Mix filaments (green symbols) over a longer time scale. No scaling was performed. Note the jump to about 35% intensity in the first two frames. (C) Steady-state light-scattering intensities plotted as a function of ParM concentration. ParM-GTP (red) and ParM-ATP (blue). The intersection of the linearly approximated curves with the x-axis defines the critical concentration. The light-scattering intensities of ParM-GTP were taken at its highest point before it started to decline. In the case of ParM-ATP filaments, the ATP was titrated slowly into the ParM solution, to avoid any turbidity and bundle formation, which happens upon rapid titration of ATP as can be seen in the stopped-flow experiments, where the ParM-ATP light-scattering signal was usually two times higher than the signal from ParM-GTP or ParM-Mix filaments. With slow titration of ATP, light-scattering intensities of ParM-ATP filaments at steady state and of ParM-GTP filaments at the highest steady-state point before declining were comparable. (D) Measuring phosphate release of 6 μM ParM upon addition of GTP (blue), ATP (red), a 1: 1 mixture of GTP and ATP (green) and a mixture of (3:1 of ATP:GTP, black). Note that even in a mixture, three parts of ATP and only one part of GTP is present; the GTPase is still predominant. The total amount of nucleotide was 5 mM.
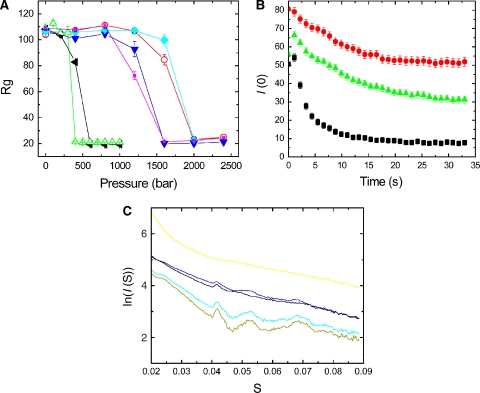
To further quantify the stability of ParM filaments with different nucleotides bound, we performed high-pressure SAX studies. Exerting pressure on a filamentous structure can lead to the destruction of noncovalent bonds, loss of nucleotide and subsequently to filament depolymerization, as shown in actin filaments (Ikeuchi et al, 2002). ParM-GTP filaments were most unstable and disintegrated earlier than ParM-ATP filaments, with the ParM-Mix filaments again titrating in between (Figure 3A and B; Supplementary movies 5 and 6). ParM polymerized with nonhydrolysable nucleotides were very stable and disintegrated at substantially higher pressures (Figure 3A). The instability or disintegration of ParM-ATP or ParM-GTP filaments is caused by nucleotide release, as we will show later. In either case, disintegration of ParM filaments was complete at relatively low pressures of about 400 bar and directly reflected how tightly the two nucleotides (ADP and GDP) were bound in the filamentous state.
Figure 3.
(A) The radii of gyration of the complexes obtained from SAX patterns are plotted as a function of static pressure. ParM-GTP (green) and ParM-ATP (black) depolymerize at pressures of 400 and 500 bar, respectively, whereas ParM-GTPγS (pink), ParM-GMPPNP (blue), ParM-ATPγS (cyan), and ParM-AMPPNP (orange) disintegrate at much higher pressures. The protein concentration was 10 mg/ml (B) Following a rapid pressure jump (1 → 400 bar), the forward scattering intensity was monitored over time. ParM-GTP filaments (black) depolymerize much sooner than ParM-Mix (green) and ParM-ATP filaments (red). (C) SAX patterns from ParM in the presence of various nucleotides. ParM-GMPPNP (light green) and ParM-AMPPNP filaments (cyan) have the highest polymer content, with predominant peaks noticeable. The sharp large first peak arises from the 24 Å subunit repeat. ParM-ATP (brown) and ParM-GTP filaments (blue) have less helical content and more monomers present, indicative by the weakening of the 24 Å and higher-order peaks and an increase in diffusive background. ParM even at high concentrations of 10 mg/ml did not polymerize in the presence of GDP, giving a SAX pattern typical for a monomer (yellow).
Static SAX measurements also confirmed that the content of polymer was highest for nonhydrolysable nucleotides, and less for ParM-ATP and ParM-GTP filaments at steady state, whereas ParM could not form a polymer with GDP or ADP (Garner et al, 2004) even at high ParM concentrations (Figure 3C).
We also monitored the ATPase and GTPase activity of ParM upon polymerization by measuring phosphate release. After adding GTP, we observed extremely rapid hydrolysis of GTP (GTP burst), which lagged behind filament polymerization, whereas ATP hydrolysis was comparably a much slower process. Even when added a 3:1 mixture of ATP to GTP, polymerization was dominated by a GTP burst (Figure 2D). This behaviour is entirely different from actin and resembles that of G-proteins, which act as molecular switches, being active only when bound to GTP and inactive upon hydrolysis of GTP to GDP (Bourne et al, 1991). The continued slow ATP hydrolysis, opposed to the complete halt of GTPase activity at steady state, already indicates why ParM-ATP filaments may be rescued after a catastrophe, whereas ParM-GTP filaments disintegrate to completion. In our present in vitro study, we have used nucleotide concentrations of either 5 or 10 mM, which is higher than that found in the cell to avoid poisoning effects from released GDP or ADP molecules. Yet we did some controls in our stopped-flow experiments with only 1 mM GTP present. The results indicated that there was no substantial difference between 5 and 1 mM of nucleotide, and the depolymerization rate was similar in both cases (see Supplementary Figure S9). These controls implied that the conclusions drawn from nucleotide concentrations of 5–10 mM may not be very different from those obtained at lower concentrations of 1–3 mM, as found in the cell.
The structure of ParM
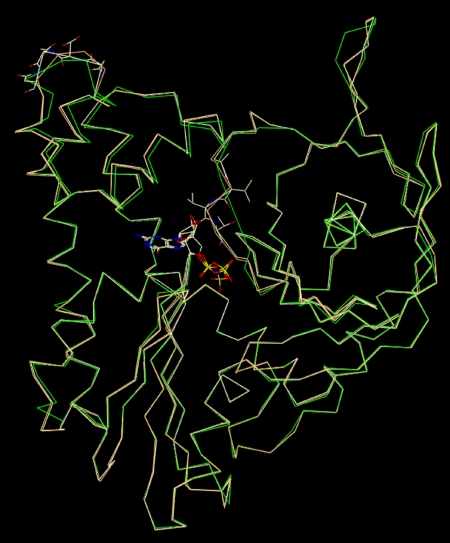
To see whether various nucleotides bind differently or cause conformational changes of ParM, we crystallized the monomer in the presence of GDP and GMPPNP and compared these structures with the known atomic structure of ParM-ADP (van den Ent et al, 2002). GDP- and GMPPNP-bound ParM crystals grew to form the same space group as ParM-ADP crystals (tetragonal, P41) (van den Ent et al, 2002) and their structures were solved by molecular replacement. A comparison of both GDP-ParM and GMPPNP-ParM structures (2.25 Å resolution) with the ParM-ADP structure (van den Ent et al, 2002) revealed that all nucleotides bind at the same site, at the interdomain cleft between subunits I and II, in a highly conserved region of residues, known as actin fold (Bork et al, 1992; Figure 4). Overlaying all three structures showed that there are only minor differences between them (Figure 4).
Figure 4.
Overlay of three different crystal structures of ParM. The already known ParM-ADP structure (green) (van den Ent et al, 2002) and our structures of ParM-GMPPNP (cream) and ParM-GDP (orange). The structures are very similar with slight variations in the conformations of some loops. All nucleotides (in CPK colour coding in the centre of the molecule) bind to the actin fold between domains I and II.
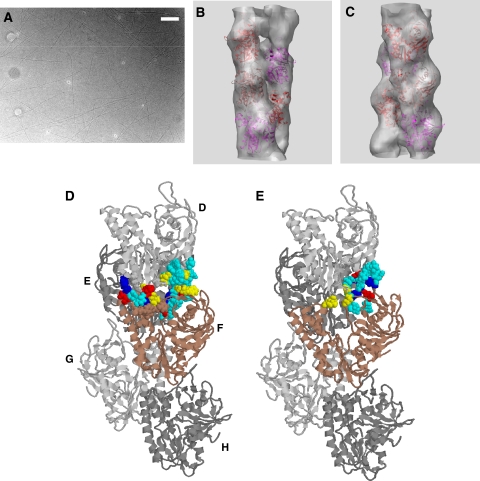
We used a combination of helical reconstruction (EOS, Yasunaga and Wakabayashi, 1996) and single-particle analysis to obtain the 3D structure of negatively stained ParM-GMPPNP filaments (Figure 5A). The general outline of the procedures that we used to calculate the ParM structure from the electron micrographs is described in detail in the Supplementary data. However, the information on the handedness of the ParM helix is lost in any EM image, and hence in any subsequent 3D reconstruction (Belnap et al, 1997). Tilt experiments have been used to determine the handedness of biological macromolecules imaged by EM (Klug and Finch, 1968; Finch, 1972). By comparing two sets of the same ParM filaments, untilted and tilted by 30° along their axis, similar to the one described by Belnap et al (1997), we found ParM to form left-handed double helical filaments, opposed to the well-characterized right-handed actin filaments (Holmes et al, 1990; Figure 5B–D). The exact procedure how we determined the handedness is described in detail in the Supplementary Figures 4S, 5S and Table S1. We determined the helical parameters to be 37 ParM molecules per 13 turns with a repeat of about 300 Å (see the power spectrum in Supplementary Figure S1). Then we fitted the atomic crystal structure of ParM-ADP into the (23 Å) electron-density map (see the Fourier Shell Correlation in Supplementary Figure S3) using a search that placed the ParM monomer into the ParM helix in all possible orientations using six parameters, three rotational and three translational. One orientation matched the observed electron density significantly better than all other solutions and this best fit was further refined by energy minimization (Figure 5B and C; Supplementary Figure S6). Docking and energy minimization procedures are described in detail in the Supplementary data. In our model, the large domain (comprising subdomain Ia and Ib; van den Ent et al, 2002) is at small radius, and the small domain (subdomains IIa, IIb; van den Ent et al, 2002) is at large radius (Figure 5D). The maximum diameter is between 80 and 85 Å. The long α-helices H10 and H12 of subdomain I (van den Ent et al, 2002) are arranged almost parallel to the filament axis. The large domain is about 48 Å in its long dimension and fits rather naturally along the long left-handed two-start ParM helix. The C-terminus is at largest radius (≈40 Å). Our atomic model of the ParM-GMPPNP filament allows us to identify the residues involved in intra- and interstrand interactions between protomers. Intrastrand interactions are extensive, involving many polar and electrostatic interactions: 13 pairs of hydrogen bonds (107–42, 108–42, 113–38, 116–40, 237–161, 238–161, 238–164, 297–240, 297–241, 33–111, 39–114, 43–112 and 43–113) and 4 salt bridges (D164–R238, D161–R238, E35–R110 and D241–R297). The interstrand interactions are also polar: six pairs of hydrogen bonds (110–254, 258–35, 258–54, 262–63, 34–261 and 34–265) and three salt bridges (E35–K258, D63–R262 and E254–R110).
Figure 5.
(A) A typical electron micrograph of negatively stained ParM-GMPPNP filaments. Scale bar, 200 nm. (B, C) Two different views of the 3D electron-density map of ParM-GMPPNP filaments, obtained from micrographs as shown in (A), with a cutoff at the contour enveloping 130% volume, is superimposed by the ribbon model of the ParM-ADP monomer crystal structure (closed conformation) orientated to give the optimal fit. (D). The best atomic model of the ParM-GMPPNP filament. In all models shown, the pointed end of the filament is at the top (subdomains Ib and IIb) and the barbed end at the bottom (subdomains Ia and IIa) (Holmes et al, 1990). We show the main interactions between subunit F and its neighbouring monomers E and D in the ParM helix. Red and blue represent salt bridges, cyan are hydrogen bonds and yellow hydrophobic residues. (E) Testing how nucleotide release effects intra- and interstrand interactions. The nucleotide-free crystal structure in the open conformation (van den Ent et al, 2002) was fitted into the electron-density map and energy minimization was performed. Our model is shown in (D). All molecules are in the closed conformation except monomer F (brown colour coding), which is in the nucleotide-free open conformation. Note that various interactions to neighbouring monomers D and E are broken when upon loss of nucleotide the cleft between subdomain I and II of monomer F opens. Interactions between the other neighbouring monomers G and H are not shown in detail.
We further elucidated the correctness of our model, using oriented gels. ParM-GMPPNP filaments like f-actin can be oriented in capillary tubes (Popp et al, 1987; Yamashita et al, 1998), and the resulting oriented gels (see Supplementary Figure S7) gives detailed X-ray diffraction patterns that diffract to high resolution. A comparison between the X-ray diffraction pattern of f-actin and ParM filaments at 8-Å resolution revealed many differences in the layer line spacing and profiles (Figure 6A). We calculated the fibre diffraction pattern from the atomic coordinates of our EM model, taking into account the contrast variation (Holmes et al, 1990). The overall fit to the X-ray fibre diffraction was very good: the intensities of the strong low-order layer lines (1, 2, 5 and 6) were well accounted. At high resolution, peak intensities were not all accurately predicted, but practically in each case, an observed peak matched a calculated peak (Figure 6B). This indicates that our EM model for the ParM-GMPPNP filament is correct within the limits of our rigid body refinement. Oriented gels of ParM diffract to very high resolution (1.5 Å resolution, data not shown), which may provide a more detailed atomic model of the ParM filament in the future by allowing individual domain movement.
Figure 6.
(A) X-ray diffraction patterns of oriented gels of ParM-GMPPNP filaments (right half) and f-actin (left half). Note the difference in the positions and intensity profiles of the layer lines. (B) The experimental X-ray diffraction pattern from ParM-GMPPNP fibres (left half) is compared with the simulated diffraction pattern from our EM model (right half). The resolution at the edge was about 8 Å. The R-factor was 0.47. The R-factor was defined as R=∑∣Iobs−k*Icalc∣/∑Iobs, with the scaling factor k*=Icalc2/Iobs Icalc. (C) The experimental X-ray diffraction pattern from ParM-GMPPNP fibres (left half) is compared with the simulated diffraction pattern from the Egelman model (Orlova et al, 2007, protein data bank code 2QU4.pdb, right half). The R-factor of the Egelman model was 0.68 and thus much worse than our EM model of ParM. Note that the characteristic near equatorial double horseshoe pattern (arrow 1) from oriented gels of ParM-GMPPNP filaments (left half) cannot be reproduced by the Egelman model. Other strong reflections produced by the Egelman model are not accounted for in the experimental fibre diffraction data (three examples are shown by the arrows on the right half) and other reflections experimentally observed (examples are shown by arrows on the left side) are missing in the diffraction of the Egelman model. Also note the difference in the layer line spacing between the left and right hand side.
Discussion
How is physiological function of ParM related to its atomic structure? As we have shown, ParM will preferably bind to GTP rather than ATP, both nucleotides being present in millimolar concentrations inside a bacterium (Schneider and Gourse, 2004).
It was shown that both in vivo (Møller-Jensen et al, 2002) and by reconstituting the ParM–ParR–parC complex in vitro (Garner et al, 2007), a bidirectional bundle of several ParM filaments formed upon the initial binding of the ParR–parC complex to ParM and that ParM polymerization pushed the plasmids to opposing poles of the bacterium (Garner et al, 2007). Here, we demonstrate that in contrast to the previous notion that ParM is a purely ATP-driven ‘polymerization motor' (Garner et al, 2004), ParM is a predominantly GTP-driven molecular switch. Biphasic polymerization observed with GTP is much more rapid than with ATP and is accompanied by a GTP burst. The ATPase activity was a comparably slow process.
Phosphate release lagged the polymerization rate for both ParM-GTP and ParM-ATP filaments. This behaviour is similar as reported in actin filaments, in which after adding actin monomers to the growing filament end as ATP-actin, ATP is rapidly hydrolysed to ADP and inorganic phosphate (Pi), both remaining attached to the filament host and only after a delay Pi is released into solution, generating ADP-actin (Carlier and Pantaloni, 1986; Carlier, 1987). At steady state, ParM filaments show nucleotide-dependant dynamic instability. Although ParM-ATP filaments have the tendency to switch spontaneously between phases of steady elongation and rapid shortening, similar to microtubules, ParM-GTP filaments cannot be rescued after reaching steady state and desintegrate steadily to completion. This intrinsic mechanism of the ParM-GTP filament is important to ensure even redistribution of ParM molecules after successful plasmid segregation in mother and daughter cell before cell division.
Based on our results, we propose a simple picture of the events involved in bacterial plasmid segregation in vivo. First, other unknown factors like clock proteins may trigger the initial binding of ParR–parC complex to ParM. This would be followed by rapid GTP-driven bidirectional polymerization of a ParM bundle formed spontaneously due to molecular crowding pushing the plasmids apart. For E. coli, with a size of about 3 μm and a ParM concentration around 10 μM (Møller-Jensen et al, 2002), it would take only about 15 s for the plasmids to be segregated and hit opposing cell walls, the impact and force from the membrane leading to unbinding of the ParR–parC complex. Now all ParM monomers in the filament bundle would be in the inactive GDP state. GDP would be released only from the monomers at filament ends, as these are freely exposed to the solvent. Upon loss of nucleotide, these monomers undergo a large conformational change (from closed to open), as described in the literature (van den Ent et al, 2002), leading to the dissociation of these subunits, as we describe in more detail below.
This view is supported by our in vitro experiments and our EM model. First, we would like to point out a major difference between actin and ParM. Whereas monomeric ParM is a completely stable protein without any bound nucleotide, actin unfolds and irreversibly denatures upon loss of its nucleotide. Therefore, in the f-actin filament under physiological conditions, its major nucleotide ADP stays bound, whereas in the ParM filament, the loss of nucleotide can happen more readily and will not have any drastic effects such as protein unfolding. This interpretation is directly reflected by our high-pressure SAX studies (Figure 3A and B). Whereas f-actin depolymerizes at very high pressures above 2000 bar (Ikeuchi et al, 2002 and our own results, data not shown), ParM-GTP and ParM-ATP filaments are depolymerized at much lower pressures of 400 and 500 bar, respectively. These results directly indicate how tightly the nucleotides are bound. In f-actin, it is extremely tightly bound, as actin would denature otherwise. In the ParM filament, the nucleotide is much less tightly bound than in f-actin and GDP is lost more readily than ADP, as can be seen from the kinetics of our pressure jump measurements (Figure 3B).
We have tested how the release of GDP would effect in- and interstrand interactions by placing the nucleotide-free ParM crystal structure into the EM electron-density map and again performing energy minimization (Figure 5E). We find that several vital interactions are lost, compared with the nucleotide-bound conformation. In the nucleotide-free filament model in-strand interactions are reduced from four to two salt bridges between domains F–D and F–H, respectively. The number of hydrogen bonds formed drops from 13 with nucleotide to only 6 for in-strand contacts between monomers F–D. They involve the following residues: 110–35, 115–40, 116–42, 43–113, 43–114 and 61–188. Between monomers F and H, 10 pairs of hydrogen bonds remain in the nucleotide-free state, which is lower by 3 from the nucleotide-bound state. Interstrand interactions are also reduced upon loss of nucleotide. The three salt bridges formed between monomers F–E and F–G in the nucleotide-bound state are reduced to two for F–E interaction (residues 208–110 and 35–258) and only one salt bridge (35–258) remains for the F–G interaction. The number of hydrogen bonds also drops from six pairs in the nucleotide-bound conformation to three in the nucleotide-free conformation.
The reduction of interactions upon loss of nucleotide thus leads to depolymerization and the dynamic instability observed. Following the actin terminology, ParM monomers have a higher probability to disintegrate from the barbed end (Holmes et al, 1990; Figure 5E), as more interactions are lost at this side of the filament compared with the pointed end.
Like microtubules, both ParM-ATP and ParM-GTP filaments face a catastrophe when their ATP or GTP cap is lost. We propose a simple explanation for the observation that ParM-ATP filaments can be rescued after a catastrophe occurs, whereas ParM-GTP filaments cannot. As outlined above, ParM polymers will start to depolymerize when the nucleotide (GDP or ADP) is released from the monomers at the ends of the filament. These monomers at the filament ends will undergo a conformational change from the closed configuration (nucleotide bound) within the ParM helix into the open conformation (nucleotide-free) and then disintegrate from the filament. Under these circumstances, a rescue of the filament is impossible as ParM-GTP or ParM-ATP monomers, which are in the closed conformation, cannot bind to monomers in the open conformation due to steric hindrance. Whether the rescue can take place depends on how fast the nucleotides GDP and ADP are released from the monomers at filament ends, causing these monomers to switch their conformation from closed to open. Our high-pressure SAX experiments showed that GDP is released faster than ADP. Our TIRF microscopy results on nucleotide-dependent steady-state behaviour indicate that in the case of ParM-GTP filaments at steady state, GDP is released continuously and fast enough to ensure that monomers at filament ends are always in the open conformation, making the rescue impossible. In the case of ParM-ADP filaments, ADP release is slower, giving the new ParM-ATP monomers from solution a chance to bind occasionally, when the monomer at the filament end is still in the closed conformation. In this case, the polymer will start to elongate again. This type of rescue is a stochastic process, as observed in our in vitro TIRF experiments. The fact that ParM-ATP filaments can be rescued after a catastrophe, whereas ParM-GTP filaments cannot, is also reflected by the behaviour of the ATPase and GTPase activity. After the first initial burst, the GTPase activity stops, whereas the ATPase is a continued slow process (Figure 2D).
Disintegration is observed only at the ends of the unidirectional ParM bundles typically observed in vivo due to molecular crowding (Møller-Jensen et al, 2002) and in vitro (Popp et al, 2007c), as short-range filament interactions within the bundle hinder both nucleotide release and filament breakage and thus disintegration. This notion is supported by our electron micrographs, which show that both ParM-ATP and ParM-GTP bundles are spindle-shaped at steady state (Figure 1C) in contrast to ParM bundles with nonhydrolysable nucleotides, which are uniform in length with blunted ends (Popp et al, 2007a).
ParM-Mix filaments behave in manner similar to ParM-GTP filaments, the depolymerization rate being slower. Nevertheless, both ParM-GTP and ParM-Mix filaments, unlike ParM-ATP filaments, will depolymerize to completion within about 10 min over the dimensions of the bacterium (Figure 1F–J; Supplementary movies 3 and 4). This is well below the division time for E. coli, of about 20 min, thus ensuring equal ParM distribution between mother and daughter cell. As our results show that ParM binds to GTP preferentially even at ATP:GTP ratios of 3:1, it is safe to conclude that in vivo ParM filaments contain mainly bound GDP with some ADP-bound monomers sprinkled in, which will not affect the ability of the filament to disassemble to completion after segregating the ParR–parC complex.
In summary, ParM is distinct from the atomic structure of f-actin not only by forming a left-handed helix but also with respect to its filament assembly dynamics. Despite being an actin homologue, ParM is distinguished from actin by several important properties: (i) the use of GTP rather than ATP; (ii) an extremely rapid instead of slow spontaneous nucleation; (iii) symmetrical bidirectional elongation due to spontaneous bundle formation in a crowded environment (Popp et al, 2007a, 2007b, 2007c) and (iv) dynamic instability as a GTP-driven molecular switch resulting in a phase of steady elongation followed by a phase of shortening to complete disintegration of the polymer instead of steady-state treadmilling.
Recently, a model for the ParM filament based on both negatively stained and cryo-EM ParM-AMPPNP filaments has been proposed by the Egelman group (Orlova et al, 2007). As in our model of the ParM-GMPPNP filament, the Egelman model shows ParM to be a left-handed helix opposed to the right-handed actin helix. Yet otherwise, the two models are in disagreement. For example, in our model, the ParM monomers within the ParM filament are in the closed conformation, whereas in the Egelman model they are in the open configuration. We have tested the correctness of the Egelman ParM structure by calculating the diffraction pattern from their model (protein data bank code 2QU4.pdb), taking into account the contrast variation (Holmes et al, 1990). Then we compared this to the experimentally observed X-ray fibre diffraction pattern from oriented gels of ParM-GMPPNP. Note that the X-ray fibre diffraction patterns from oriented gels of ParM-GMPPNP and ParM-AMPPNP are virtually identical (Supplementary Figure S8), indicating that there is no major structural difference between ParM-AMPPNP and ParM-GMPPNP filaments. We find that the Egelman model does not fit to our experimentally observed X-ray fibre diffraction pattern well (Figure 6C). The Egelman model produces layer lines that are not observed experimentally, whereas other strong layer lines visible in the X-ray diffraction pattern of the oriented ParM gels are missing. The characteristic double horseshoe-like pattern near the equator observed from oriented ParM fibres also cannot be accounted for by the Egelman model. Furthermore, the helical parameters of the Egelman model differ from the parameters we observe both from X-ray diffraction patterns of oriented gels and from negatively stained ParM-GMPPNP filaments. In the Egelman model, helical parameters are assigned as l=14n+31m, whereas we find the helical parameters to be l=17n+37m. The R-factor of the Egelman model against the experimentally observed fibre diffraction data is 0.68, whereas our EM model even with the limitation of rigid body refinement is much better at 0.47. These findings strongly argue in favour of our model of the ParM filament.
Still remains one intriguing puzzle in bacteria: so far, no motor-, filament-severing- or formin-like proteins that are prominent components of the eukaryotic cytoskeleton and that control the dynamic behaviour of actin filaments have been discovered. Our hypothesis is that they simply do not exist and bacterial cells manage with ‘fibrous protein polymerization motors', which work by a well-defined balance between nucleotide-driven nonequilibrium polymerization and dynamic instability. Whether a filament shows dynamic microtubule-like instability, disintegrates to completion or treadmills or neither (if both ends have similar rate constants) seems to depend rather critically on a balance of all various binding constants and is not specific to particular classes of proteins.
Materials and methods
TIRF microscopy and biochemical experiments
Single-molecule visualization of polymerization and steady-state behaviour of ParM filaments upon addition of various nucleotides were undertaken similarly as described previously (Popp et al, 2007c). Buffer conditions, crowding agents, labelling of ParM with fluorophores and the optical setup using an inverted Nikon 2000 microscope equipped with perfect focus and subsequent data analysis fitting the filament length with a polygon by hand using Image J, Aqua Cosmos and Kaleidograph were similar as described in detail previously (Popp et al, 2007c). Initially, as in our previous experiments (Popp et al, 2007c), we added 10 mM of nucleotide to ParM to initiate polymerization. Yet we found that the polymerization rates and steady-state behaviour did not change when lowering the nucleotide concentration to 5 mM, so subsequently, we mostly used this concentration, as it is closer to the physiological nucleotide concentration within the cell (about 1–3 mM).
Clean glass slides were very important to obtain reproducible results. We tried various procedures of cleaning as described by other groups to make the surface of the glass hydrophilic (Kuhn and Pollard, 2005), and we also tested additional procedures like blocking the surface with BSA, which increases the hydrophobicity and, in some cases, decreases the high mobility of the filaments. The results obtained from all these glass surfaces were rather similar, yet the washing procedure we mostly used (Popp et al, 2007b, 2007c) was fast and produced reliable hydrophilic surfaces. We want to point out in this context that according to the principal predictions of adsorption theory, proteins do not adsorb to hydrophilic surfaces and are separated from the surface by an aqueous layer (Vogler, 1998). We have recently proven this hypothesis to be correct for actin-containing filaments (Popp et al, 2007a, 2007b). ParM filaments behaved similarly. Therefore, for most filaments, only the total length of the filament could be studied over time, owing to the large motions the filaments underwent near the glass surface. Only for a small amount of filaments (less than 3% of all filaments) the behaviour of both filament ends could be followed at steady state. This was possible, when either a filament appeared firmly adsorbed to the surface or fluorescent speckles in the filament could be used as a ficidual mark.
Stopped-flow experiments were performed with an SF61-DX2 stopped-flow spectrophotometer (Hi-Tech Scientific, Salisbury, UK). Filament formation at 25°C was monitored using light-scattering as described previously (Bagshaw and Trentham, 1973). Software provided by Hi-Tech Scientific was used for curve fitting of the data.
Conventional light-scattering experiments on ParM polymerization was carried out at 25°C using a Hitachi Fluorescence Spectrometer F-4550 at the wavelength of 660 nm.
The release of inorganic phosphate upon nucleotide hydrolysis during ParM polymerization was measured at 25°C using Phosphate Assay Kit (E-6646) from Molecular Probes, based on a method described previously (Webb, 1992).
SAX and high-pressure SAX
ParM in P-buffer (25 mM Hepes, pH 7.5, 30 mM KCl, 2 mM MgCl2 and 5 mM DTT) at concentrations between 5 mg/ml and 15 mg/ml were polymerized by the addition of 5 or 10 mM nucleotide and placed into specially designed chambers (Fujisawa, 2000; Nishikawa et al, 2001). Temperature was controlled at 25°C. Hydrostatic pressure was applied either in multiple steps or by a single step within about 1 s (Nishikawa et al, 2001). Using beam line BL45-XU (λ=0.9 Å) and camera lengths of 1–2.2 m, the small-angle-scattering pattern of ParM was recorded on a cooled CCD camera and data were analysed as described previously (Fujisawa, 2000).
X-ray crystallography
ParM-GDP crystals were grown under conditions similarly as previously described for ParM-ADP (van den Ent et al, 2002). ParM-GMPPNP crystals were prepared by soaking GMPPNP into ParM-GDP crystals. X-ray data to a resolution of 2.25 Å were collected at BL45 and the ParM-GDP and ParM-GMPPNP structures were solved by molecular replacement.
Oriented gels
ParM-GMPPNP and ParM-AMPPNP filaments were oriented in glass capillaries as previously described for f-actin and other biological polymers (Popp et al, 1987; Yamashita et al, 1998). The cross-section of the X-ray beam at BL45-XU (λ=0.9 Å) was optimized to 150 × 150 μm. The camera length was 85 cm and fibre diffraction patterns were recorded on Rigaku imaging plates. The exposure time was 40 s per image. X-ray fibre diffraction data were analysed using the software package developed by K Hasegawa and K Namba (unpublished data), which allowed, among other features, stripping of the background, averaging of data sets or quadrants and correcting for angular disorientation.
Electron microscopy
A drop of ParM-GMPPNP filaments was applied to carbon-coated copper grids, blotted stained with 1% uranyl acetate and visualized under a Jeol JEM-2010 HC microscope operated at 100 keV and a nominal magnification of 40 000. Films were digitized with PhotoScan2000 (Z/I Imaging) at 7 μm steps. The handedness of ParM was determined by tilting a filament by 30° along its axis and comparing the two views as described previously (Belnap et al, 1997). Reconstructions of ParM were performed as described in detail recently (Narita and Maéda, 2007). Exact procedures of how the reconstruction was performed and how the handedness was determined are given in the Supplementary data.
Supplementary Material
Supplementary movie 1
Supplementary movie 2
Supplementary movie 3
Supplementary movie 4
Supplementary movie 5
Supplementary movie 6
Supplementary Figures and Tables
Supplementary data
Acknowledgments
We thank Shiho Minakata for taking the X-ray data of ParM crystals, Kazuki Ito for beam line support at Spring8 BL45-XU and Akihiro Yamamoto for technical supervision of our light microscopy facilities. ParM plasmid was a kind gift from Jakob Møller-Jensen. We are indebted to Harold P Erickson for initially pointing out to us the possible importance of GTP in ParM polymerization.
References
- Bagshaw C, Trentham D (1973) The characterization of myosin–product complexes and of product-release steps during the magnesium ion-dependent adenosine triphosphate reaction. Biochem J 141: 331–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belnap DM, Olson NH, Baker TS (1997) A method for establishing the handedness of biological macromolecules. J Struct Biol 120: 44–51 [DOI] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A (1992) An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA 89: 7290–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349: 117–127 [DOI] [PubMed] [Google Scholar]
- Carlier M-F (1987) Measurement of Pi dissociation from actin filaments following ATPase hydrolysis using a linked enzyme system. Biochem Biophys Res Commun 143: 1069–1075 [DOI] [PubMed] [Google Scholar]
- Carlier M-F, Pantaloni D (1986) Direct evidence for ADP-Pi-F-actin as the major intermediate in ATP-actin polymerization. Rate of dissociation of Pi from actin filaments. Biochemistry 35: 7789–7792 [DOI] [PubMed] [Google Scholar]
- Finch JT (1972) The hand of the helix of tobacco virus. J Mol Biol 66: 291–294 [DOI] [PubMed] [Google Scholar]
- Fujisawa T (2000) Structural studies on protein solutions by using solution X-ray scattering technique at RIKEN structural biology beamline I (BL45XU). J Cryst Soc Jpn 42: 97–105 [Google Scholar]
- Garner EC, Cambell CS, Mullins RD (2004) Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306: 1021–1025 [DOI] [PubMed] [Google Scholar]
- Garner EC, Cambell CS, Weibel DB, Mullins RD (2007) Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 315: 1270–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Møller-Jensen J, Bugge-Jensen R (2000) Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol 37: 455–466 [DOI] [PubMed] [Google Scholar]
- Holmes KC, Popp D, Gebhard W, Kabsch W (1990) Atomic model of the actin filament. Nature 347: 44–49 [DOI] [PubMed] [Google Scholar]
- Ikeuchi Y, Suzuki A, Oota T, Hagiwara K, Tatsumi R, Ito T, Balny C (2002) Fluorescence study of the high pressure-induced denaturation of skeletal muscle actin. Eur J Biochem 269: 364–371 [DOI] [PubMed] [Google Scholar]
- Klug A, Finch JT (1968) Structure of viruses of the papilloma-polyoma type: IV. Analysis of tilting experiments in the electron microscope. J Mol Biol 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD (2005) Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J 88: 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller-Jensen J, Jensen RB, Gerdes K (2000) Plasmid and chromosome segregation in prokaryotes. Trends Microbiol 8: 313–320 [DOI] [PubMed] [Google Scholar]
- Møller-Jensen J, Jensen RB, Löwe J, Gerdes K (2002) Prokaryotic DNA segregation by an actin-like filament. EMBO J 21: 3119–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita A, Maéda Y (2007) Molecular determination by electron microscopy of the actin filament end structure. J Mol Biol 365: 480–501 [DOI] [PubMed] [Google Scholar]
- Nishikawa Y, Fujisawa T, Inoko Y, Moritoki M (2001) Improvement of a high pressure cell with diamond windows for solution X-ray scattering of proteins. Nucl Instrum Methods Phys Res A 467: 1384–1387 [Google Scholar]
- Orlova A, Garner EC, Galkin VE, Heuser J, Mullins RD, Egelman EH (2007) The structure of bacterial ParM filaments. Nat Struct Mol Biol 14: 921–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp D, Lednev VV, Jahn W (1987) Methods of preparing well-orientated sols of f-actin containing filaments suitable for X-ray diffraction. J Mol Biol 197: 679–684 [DOI] [PubMed] [Google Scholar]
- Popp D, Yamamoto A, Iwasa M, Nitanai Y, Maeda Y (2007a) Single molecule polymerization, annealing and bundling dynamics of SipA induced actin filaments. Cell Motil Cytoskeleton; e-pub ahead of print December 12 [DOI] [PubMed] [Google Scholar]
- Popp D, Yamamoto A, Maeda Y (2007b) Crowded surfaces change annealing dynamics of actin filaments. J Mol Biol 368: 365–374 [DOI] [PubMed] [Google Scholar]
- Popp D, Yamamoto A, Narita A, Iwasa M, Maeda K, Maeda Y (2007c) Concerning the dynamic instability of actin homolog ParM. Biochem Biophys Res Commun 353: 109–114 [DOI] [PubMed] [Google Scholar]
- Schneider DA, Gourse RL (2004) Relationship between growth rate and ATP concentration in Escherichia coli: a bioassay for available cellular ATP. J Biol Chem 279: 8262–8268 [DOI] [PubMed] [Google Scholar]
- van den Ent F, Møller-Jensen J, Amos LA, Gerdes K, Löwe J (2002) F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J 21: 6935–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler EA (1998) Structure and reactivity of water at biomaterial surfaces. Adv Colloid Interface Sci 74: 69–117 [DOI] [PubMed] [Google Scholar]
- Webb MR (1992) A continous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci USA 89: 4884–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I, Suzuki H, Namba K (1998) Multiple-step method for making exceptionally well oriented liquid-crystalline sols of macromolecular assemblies. J Mol Biol 278: 609–615 [DOI] [PubMed] [Google Scholar]
- Yasunaga T, Wakabayashi T (1996) Extensible and object-oriented system Eos supplies a new environment for image analysis of electron micrographs of macromolecules. J Struc Biol 116: 155–160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary movie 1
Supplementary movie 2
Supplementary movie 3
Supplementary movie 4
Supplementary movie 5
Supplementary movie 6
Supplementary Figures and Tables
Supplementary data