Abstract
Cystatins are a family of naturally occurring cysteine protease inhibitors, yet the target proteases and biological processes they regulate are poorly understood. Cystatin F is expressed selectively in immune cells and is the only cystatin to be synthesised as an inactive disulphide-linked dimeric precursor. Here, we show that a major target of cystatin F in different immune cell types is the aminopeptidase cathepsin C, which regulates the activation of effector serine proteases in T cells, natural killer cells, neutrophils and mast cells. Surprisingly, recombinant cystatin F was unable to inhibit cathepsin C in vitro even though overexpression of cystatin F suppressed cellular cathepsin C activity. We predicted, using structural models, that an N-terminal processing event would be necessary before cystatin F can engage cathepsin C and we show that the intracellular form of cystatin F indeed has a precise N-terminal truncation that creates a cathepsin C inhibitor. Thus, cystatin F is a latent protease inhibitor itself regulated by proteolysis in the endocytic pathway. By targeting cathepsin C, it may regulate diverse immune cell effector functions.
Keywords: cathepsin, cystatin, lymphocytes, proteolysis
Introduction
The cystatins are naturally occurring cysteine protease inhibitors found either within the cytosol or secreted from cells (Abrahamson et al, 2003; Kopitar-Jerala, 2006). In humans, there are currently 11 family members divided into three subgroups depending on the presence of single or multiple ‘cystatin' domains and the presence or absence of a signal sequence. Cystatins are potent inhibitors of the C1 family of cysteine proteases and some members of the type II subgroup also inhibit the C13 family protease asparaginyl endopeptidase (AEP)/legumain (Alvarez-Fernandez et al, 1999; Manoury et al, 2001). The C1 cysteine proteases include the endosomal/lysosomal cathepsins, which are involved in many key biological processes such as bone remodelling as well as development and function of the immune system. These enzymes have also been linked to tumour cell invasion (Jedeszko and Sloane, 2004) and to arterial remodelling and atherogenesis (Liu et al, 2004), implying that their regulation is important. In principle, cystatins can provide this regulation but the physiological situations in which they do so are not clear. The importance of cystatins is underlined by pathological conditions that arise upon loss or mutation of some cystatin genes. For example, mice lacking cystatin M exhibit abnormal and eventually lethal defects in the development of the epidermis (Zeeuwen et al, 2002).
Several cystatin structures have now been solved, two in complex with proteases. These structures reveal the ‘cystatin fold', a five-stranded β-sheet wrapped around an extended helix and a protease-interacting ‘edge' made up of the N-terminal region and two loops found at the end of the antiparallel β-sheet (Bode et al, 1988; Stubbs et al, 1990; Jenko et al, 2003). Inhibition of AEP is due to a second distinct protease-binding site (Alvarez-Fernandez et al, 1999). Although some cystatins are partially localised within the vacuolar system of mammalian cells, type II cystatins are primarily secreted into the external milieu where they are proposed to ‘mop up' potentially harmful cysteine proteases released from cells (Abrahamson et al, 2003; Kopitar-Jerala, 2006).
Cystatin F is a type II cystatin whose expression is limited primarily to cells of the immune system such as T cells, natural killer (NK) cells and dendritic cells (Halfon et al, 1998; Ni et al, 1998; Hashimoto et al, 2000; Obata-Onai et al, 2002). Cystatin F was identified as one of the most upregulated transcripts in monocyte-derived dendritic cells undergoing LPS-induced maturation (Hashimoto et al, 2000) and was independently identified as CMAP (cystatin-like metastasis associated protein) whose level of expression correlated with metastatic potential in liver tumours (Morita et al, 1999). Cystatin F has relatively low sequence homology to other family members (∼35%), the main distinguishing features being an extended N-terminal region, two additional cysteine residues and non-conservative substitutions in the putative protease-interacting domains. Cystatin F was shown to be secreted as a disulphide-linked dimer (Cappello et al, 2004), which is inactive until it is reduced to its monomeric form (Langerholc et al, 2005). This form was shown to inhibit cathepsins L, V, K and F most potently, while cathepsins S and H were less sensitive and cathepsins B and C were not inhibited (Halfon et al, 1998; Ni et al, 1998; Langerholc et al, 2005).
We recently described the crystal structure of human cystatin F. Its dimeric form is stabilised by two inter-subunit disulphide bridges between Cys26 in the extended N terminus of one monomer and Cys63 on the other monomer (Schuettelkopf et al, 2006). The resulting dimer is unable to bind to C1 family cysteine proteases, due to mutual steric hindrance of the protease-binding sites. Cystatin F is the only cystatin to be made as an inactive precursor, indicating that its activity can be regulated and suggesting an intracellular function. Indeed, compared with cystatin C, a much larger fraction of cystatin F in U937 cells is directed to intracellular compartments (Nathanson et al, 2002).
We reasoned that the target proteases and potential functions of cystatin F in immune cells might be elucidated by isolation of endogenous cystatin F–protease complexes. Surprisingly, given earlier data that cathepsin C could not be inhibited by cystatin F (Langerholc et al, 2005), we show that this enzyme is one of its principal interacting partners. We resolve this anomaly by showing that cystatin F must undergo an N-terminal processing event to acquire cathepsin C inhibitory capacity. Since cathepsin C/DPPI is essential for the activation of a range of granule-localised serine proteases in T cells, NK cells, neutrophils and mast cells, cystatin F may have an important regulatory role in immune cells.
Results
Cystatin F expression in immune cells
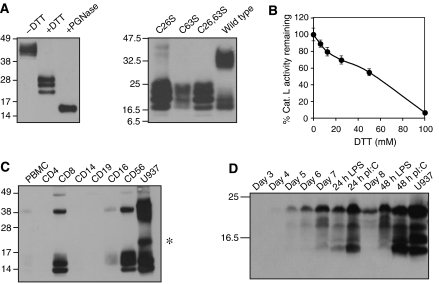
To study cystatin F, we expressed it using a vector that permitted substantial amounts to be secreted from CHO cells (Li et al, 2003). As expected the purified protein was a disulphide-linked dimer and showed several distinct forms due to heterogeneous N-linked glycosylation (Figure 1A, left; Ni et al, 1998; Nathanson et al, 2002; Cappello et al, 2004). Site-directed mutagenesis of either Cys26 or Cys63 prevented dimer formation, confirming that inter-subunit disulphide bridges involved these residues (Schuettelkopf et al, 2006; Figure 1A, right). As recently shown by others (Langerholc et al, 2005), inhibitory activity against cathepsin L was only revealed following incubation with millimolar levels of reducing agents (Figure 1B). Cystatin F was expressed primarily in CD8+ T cells and in CD56+ NK cells consistent with mRNA data (Halfon et al, 1998; Obata-Onai et al, 2002) and while most appeared to be monomeric a significant amount of inactive dimer was also present (Figure 1C). Lower levels of cystatin F were observed in CD16+ cells but expression was not detected in CD4+ T cells, CD19+ B cells or in resting CD14+ monocytes. However, cystatin F expression was detected as monocytes differentiated into dendritic cells and consistent with the SAGE analysis of Hashimoto et al (2000), dendritic cell maturation with TLR ligands further increased cystatin F expression (Figure 1D).
Figure 1.
Cystatin F production and endogenous expression in immune cells. (A) Left panel: purified recombinant cystatin F is a disulphide-linked dimer with heterogeneous N-linked glycosylation. Right panel: mutagenesis of either Cys26 or Cys63 prevents dimer formation in transiently transfected 293T cells. (B) Cathepsin L inhibitory activity appears upon dimer reduction. (C) Cystatin F is expressed principally in human CD8+ T cells and in CD56+ cells, which are predominantly NK cells. In addition to dimer and monomer, an additional species (*) is seen in U937. Equal protein was loaded for all cells. (D) Cystatin F is induced as CD14+ monocytes differentiate into dendritic cells and is further induced by stimulation with TLR4 (LPS) or TLR3 (poly I:C) ligands. All gels were non-reducing except (D).
Cystatin F is complexed with cathepsin C in immune cells
To identify protease targets of cystatin F, we isolated cystatin F from detergent lysates of the human monocytic and NK cell lines U937 and YT. Parallel lysates were mixed with Sepharose beads carrying either affinity-purified anti-cystatin F antibodies or control rabbit IgG. Bound proteins were eluted and separated by SDS–PAGE. As shown in Figure 2A, several species were specifically recovered in the anti-cystatin F precipitates. MALDI TOF/TOF mass fingerprinting identified cystatin F itself and a smaller protein with apparent mol. wt. ∼7 kDa that was consistently observed in both U937 and YT samples (bands 3 and 5 in Figure 2A). This protein was identified as the light chain of cathepsin C (Supplementary Figure S1). The heavy chain of cathepsin C was also identified in the anti-cystatin F, but not in control Ig, precipitations from U937 cells (Figure 2A, band 1 and Supplementary Figure S1). Although some other cysteine proteases were also identified, we were intrigued by the association with cathepsin C, which was reported to be resistant to inhibition by recombinant cystatin F (Langerholc et al, 2005).
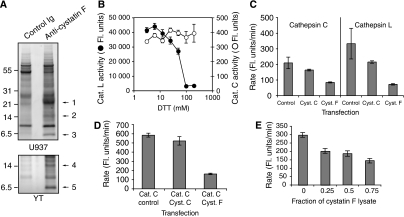
Figure 2.
Cystatin F is associated with cathepsin C in U937 and YT cells and inhibits its activity in vivo but not in vitro. (A) Proteins associated with anti-cystatin F or control antibodies were isolated from U937 and YT cells and separated by reducing SDS–PAGE. The numbered bands were excised and identified by tryptic mass fingerprinting as cathepsin C heavy chain/cystatin F (band 1), cystatin F (bands 2 and 4) and cathepsin C light chain (bands 3 and 5). Tryptic peptide coverage and peptides confirmed by MS/MS sequencing are shown in Supplementary Figure S1. (B) Cystatin F monomer generated with DTT inhibits cathepsin L (closed circles) but not cathepsin C (open circles). (C) Cystatin F inhibits cathepsin C activity in living cells; 293T cells were transfected with pcDNA3.1 DNA (empty vector control) or pcDNA3.1 into which either cystatin C or F had been cloned. After 48 h, post-nuclear cell lysates were tested for cathepsin L or cathepsin C activity. (D) As in (C) except cathepsin C was additionally expressed in 293T cells with or without cystatin C or F. (E) Lysates of 293T cells separately transfected with cystatin F or control empty vector were mixed with lysates from cathepsin C-transfected cells. The ratio of cystatin F lysate to control lysate was varied as shown and cathepsin C activity assayed as before.
Cystatin F fails to inhibit cathepsin C in vitro but suppresses its activity in vivo
We reinvestigated the capacity of cystatin F to inhibit cathepsin C in vitro. Consistent with the study of Langerholc et al (2005), reduction of cystatin F allowed inhibition of cathepsin L but not cathepsin C (Figure 2B). To investigate why cystatin F associates with cathepsin C in living cells yet cannot inhibit the enzyme in vitro, we asked whether cystatin F can inhibit cathepsin C in vivo. 293T cells were transfected with either cystatin F, cystatin C or control vector and 48 h later the activity of cathepsins L and C was measured in cell lysates using appropriate substrates. Transfection with cystatin F suppressed the activity of endogenous cathepsins L and C, as did cystatin C (Figure 2C), albeit less potently presumably because a greater fraction is secreted (Nathanson et al, 2002). Cystatin F also inhibited overexpressed cathepsin C (Figure 2D). Cystatin F might suppress cathepsin C activity directly or indirectly since, unlike most C1 cysteine proteases, cathepsin C does not autoactivate but rather is activated by other proteases (Dahl et al, 2001; Mallen-St Clair et al, 2006). We tested the possibility that cystatin F might suppress cathepsin C activation by transfecting cystatin F, cathepsin C or control vector DNA into separate populations of 293T cells and then mixing the lysates in different ratios before measuring cathepsin C activity. However, consistent with direct inhibition of activated cathepsin C, lysates from cystatin F-transfected cells were able to suppress cathepsin C activity in parallel cell lysates (Figure 2E). Inhibition was not complete, presumably because of the large dilution of inhibitor and target protease in the cell lysates.
The N terminus of cystatin F cannot be accommodated in the active site of cathepsin C
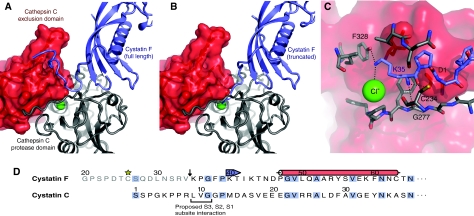
The above results presented a paradox. Cystatin F suppressed cathepsin C activity following expression in 293T cells and interacted with cathepsin C in U937, YT and 293T cells (Figure 2 and data not shown), yet recombinant monomeric cystatin F was inactive in vitro as a cathepsin C inhibitor. To try to resolve this discrepancy, we generated model structures for complexes between cystatin F and cathepsin C as well as other C1 cysteine proteases using the existing co-crystal structures of papain and stefin B (Stubbs et al, 1990), cathepsin H and stefin A (Jenko et al, 2003) and cathepsin C (Turk et al, 2001) as templates. In cystatin C and chicken egg white cystatin, the N-terminal region is an important determinant of the affinity of the inhibitor for its target proteases (Abrahamson et al, 1991; Lindahl et al, 1992). Modelling experiments showed that the extended N terminus of cystatin F could be accommodated in the cathepsin L active site, and in the active sites of other C1 family endopeptidases through placement over the non-prime subsites (S2, S3, S4, etc.) in an analogous manner to that proposed for cystatin C (Bode et al, 1990). However, this is not possible in the case of cathepsin C because the exclusion domain closes the substrate channel beyond the S2 subsite (Turk et al, 2001). As shown in Figure 3A, the cystatin F N-terminal region might fold back in a hook-like conformation similar to that observed for stefin A in complex with another aminopeptidase, cathepsin H (Jenko et al, 2003), but this is unlikely as such a conformation would bring the bulky side chain of Phe38 in cystatin F (Gly in stefin A) in close proximity with other residues, in particular Lys35. However, truncation of the cystatin F N terminus would allow it to be accommodated in the cathepsin C active site (Figure 3B). Moreover, truncation to Lys 35 might permit a favourable interaction between the Lys side chain and the chloride ion at the bottom of the S2 pocket (Figure 3C). Truncation to this point, but not beyond would align the N terminus of cystatin F with the residues known to contribute most to the binding of the N-terminal region of cystatin C to target proteases (Abrahamson et al, 1991; Figure 3D).
Figure 3.
(A) Theoretical model of the interaction of full-length cystatin F (blue) with cathepsin C (grey/red) showing the clash between the extended N terminus of cystatin F and the exclusion domain of cathepsin C. (B) Truncation of cystatin F by 15 residues removes this steric hindrance and may allow the now N-terminal Lys35 to interact with the chloride ion (green) at the bottom of the cathepsin C S2 subsite. (C) Details of the proposed interaction between truncated cystatin F and the cathepsin C S2 subsite. Possible hydrogen bonds between the amino groups of Lys35 and cathepsin C, as well as the contact between Lys35 and the chloride bound at the bottom of the S2 subsite are indicated with dotted lines. Key residues are labelled and coloured according to parent chain. (D) The N-terminal sequences of cystatin F and cystatin C are aligned and amino-acid identities are coloured in blue. The 15 amino acids removed by cleavage (arrow) in cystatin F are in grey. Cys26 participating in dimer formation in cystatin F is starred and secondary structure elements (red=α-helix, blue=β-strand) are marked above the sequence. In cystatin C, the key Leu-Val-Gly sequence for interaction with C1 cysteine protease S3-S1 non-prime sites (Bode et al, 1990) is underlined.
The cellular form of cystatin F is truncated to Lys35 to permit cathepsin C inhibition
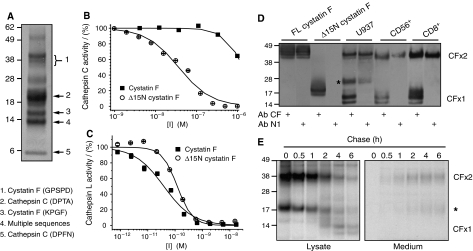
To test the prediction that cellular cystatin F undergoes N-terminal processing, we isolated cystatin F from U937 cells, separated the dimeric and monomeric forms by non-reducing SDS–PAGE and subjected them to N-terminal sequence analysis. As shown previously, U937 cells contain a mixture of dimeric inactive cystatin F and monomeric ‘active' protein (Nathanson et al, 2002; Langerholc et al, 2005; Figure 1). The dimeric form had the same N-terminal sequence (GPSPD) as secreted cystatin F, indicating that signal sequence cleavage but no further processing had occurred. Interestingly, the sequence of the monomeric form was truncated by 15 residues to lysine 35 (KPGF), exactly as suggested by the modelling analysis (Figure 4A). As expected, other bands corresponded to cathepsin C sequences (Figure 4A). We generated a new recombinant form of cystatin F lacking the first 15 residues (Δ15N cystatin F), which was secreted in monomeric form since it lacks Cys26 (Supplementary Figure S2). Crucially, whereas full-length monomeric cystatin F inhibited cathepsin C poorly, even at high concentrations (IC50>1 μM), Δ15N cystatin F was an effective cathepsin C inhibitor with an IC50 value of 34±4 nM (Figure 4B). In contrast, both full-length and truncated cystatin F were potent inhibitors of cathepsin L (Figure 4C), although the truncated form demonstrated a somewhat lower affinity for cathepsin L compared to the full-length inhibitor (IC50=120±20 versus 39±6 pM), indicating that residues N-terminal of Lys35 contribute to cystatin F binding to cathepsin L.
Figure 4.
Cellular processing of cystatin F to Lys35 generates a cathepsin C inhibitor. (A) Non-reducing SDS–PAGE separation of cystatin F immunoprecipitated from U937 cells. N-terminal sequences of cystatin F and cathepsin C are indicated. One prominent band returned multiple sequences including that of processed cystatin F. Inhibitory capacity of full-length and Δ15N cystatin F against cathepsin C (B) or cathepsin L (C) measured following reduction with DTT. (D) Lysates from U937, CD56+ and CD8+ cells were separated by non-reducing SDS–PAGE and blots were probed with antibodies against either whole cystatin F (Ab CF) or a 15-residue N-terminal peptide (Ab N1). Reactivities with recombinant cystatin F and Δ15N cystatin F are also shown. (E) Pulse-chase and non-reducing SDS–PAGE analysis of cystatin F maturation and secretion in U937 cells. Dimer (CFx2), monomer (CFx1) and an intermediate form (*) are indicated.
Thus, cystatin F only becomes a cathepsin C inhibitor following an intracellular processing event that removes the sterically hindering N terminus. To demonstrate that cystatin F is similarly processed in primary immune cells, we developed an antibody (N1) specific for the processed 15-residue N-terminal sequence. Indeed, we found that in human CD56+ NK cells and CD8+ T cells the N1 antibody reacted with inactive dimeric but not with monomeric cystatin F (Figure 4D). Metabolic pulse-chase labelling of U937 cells demonstrated that cystatin F monomer is only observed after 2–4 h of chase as the dimeric form disappears (Figure 4E). Further, while some dimer and a form migrating between dimer and monomer were secreted into the medium, the monomer was only found in cell lysates, consistent with conversion after leaving the secretory pathway (Figure 4E). The intermediate species (asterisks in Figure 4D and E) has been seen previously in U937 cells (Nathanson et al, 2002; Cappello et al, 2004). Its slower migration appears to be due to unusual glycosylation of the monomeric form since it is not affected by reduction, retains some reactivity with the N1 antibody and upon Endo H treatment, migrates similar to monomer (Supplementary Figure S3). Further characterisation of this species is needed to establish its glycosylation status and its ability to dimerise.
Cystatin F is activated following targeting to the endocytic pathway
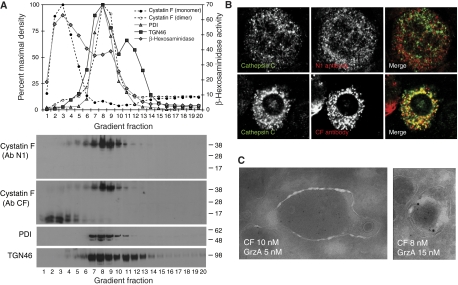
To examine the cellular localisation of dimeric and monomeric cystatin F in more detail, we disrupted U937 cells and fractionated post-nuclear supernatants on Percoll gradients. The dimeric and monomeric forms of cystatin F showed a strikingly distinct distribution. Whereas dimeric cystatin F co-migrated with markers of the ER (PDI) and Golgi apparatus (TGN46), monomeric cystatin F co-migrated with the bulk of β-hexosaminidase activity, consistent with lysosomal localisation (Figure 5A). Moreover, the latter form reacted with the antibody raised against whole cystatin F (CF) but not with the N1 antibody confirming N-terminal processing (Figure 5A). The distribution of processed and unprocessed cystatin F was also distinct in primary murine bone marrow-derived mast cells (BMMCs). In cells that expressed both cystatin F and cathepsin C, intact cystatin F, detected with the N1 antibody, was seen in many small structures some of which were positive for the Golgi marker GM130, but did not colocalise with cathepsin C (Figure 5B and Supplementary Figure S4A). In contrast, the CF antisera reactive with intact and processed cystatin F labelled many, though not all, cathepsin C-positive structures (Figure 5B). Cystatin F also partially colocalised with the cathepsin C substrate, granzyme A as well as with perforin and LAMP1 in human CD8+ T-cell blasts (Supplementary Figure S4B). Importantly, we could confirm colocalisation of cystatin F and granzyme A at the ultrastructural level in human CD8 T-cell granules (Figure 5C). Taken together, these data demonstrate conversion of dimeric to monomeric cystatin F within the endocytic pathway. Moreover, the monomeric cystatin F in dense lysosomal vesicles and those harbouring cathepsin C in murine mast cells appeared to be processed to the truncated active form.
Figure 5.
Dimeric and processed monomeric cystatin F are found in distinct cellular compartments. (A) Non-reducing SDS–PAGE analysis of Percoll gradient-fractionated U937 cells. Monomeric, processed cystatin F in U937 (detected with Ab CF) cells migrates in denser gradient fractions compared with dimeric cystatin F (detected with Abs CF and N1). ER (PDI), Golgi (TGN46) and lysosome (β-hexosaminidase) marker distributions are indicated. (B) Distinct distribution of full-length (N1 antibody reactive) endogenous cystatin F in murine BMMC. Only cystatin F lacking the N terminus (CF antibody) shows colocalisation with cathepsin C. (C) Cystatin F and granzyme A colocalise in human CD8+ T-cell blasts. Ultrathin cryosections were stained for cystatin F and granzyme A, appropriate secondary antibodies and protein A-gold as indicated.
Cystatin F and cathepsin C function in immune cells
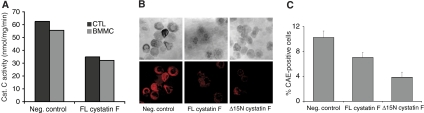
To assess directly the consequences of manipulating cystatin F levels in immune cells, we expanded primary murine T cells and mast cells from spleens and bone marrow, respectively, and transduced the cells with a bicistronic GFP-containing retroviral vector that expressed either murine wild-type cystatin F or an N-terminally truncated version analogous to the form we found in U937 cells. GFP-positive cells were sorted by FACS and cathepsin C activity was measured in cell lysates. Cathepsin C activity was significantly suppressed by cystatin F overexpression in both CD8+ T cells and mast cells (Figure 6A). In mast cells (BMMC), we also measured levels of chymase-dependent chloroacetate esterase (CAE) activity, which was reported to be completely lacking in cathepsin C-null mast cells (Wolters et al, 2001). As shown in Figure 6B and C, compared with mast cells expressing GFP alone, CAE activity was substantially suppressed by retroviral expression of cystatin F and even more so by expression of the N-terminally truncated form in spite of the fact that it was expressed at a reduced level compared with the full-length protein (Supplementary Figure S4). Taken together, these data are consistent with the idea that cystatin F can attenuate cathepsin C activity in primary immune cells.
Figure 6.
(A) Primary murine T cells (CTL) or BMMCs were infected with IRES-GFP-expressing retroviruses additionally expressing murine cystatin F, murine Δ15N cystatin F or GFP only control. GFP-positive cells were sorted by FACS and cathepsin C activity was assayed. BMMC chymase-dependent chloroacetate esterase (CAE) activity was assessed and representative images in (B) were quantitated as shown in (C).
Discussion
Cystatins have generally been thought of as ‘housekeeping' genes whose role is to inhibit inadvertently released cysteine proteases before they can cause tissue damage. In the case of cystatin F, recent data challenge this view by showing that the secreted form of cystatin F is a functionally inactive disulphide-linked dimer (Langerholc et al, 2005; Schuettelkopf et al, 2006; Figure 1B). Instead, its domain of action is likely to be within the endocytic pathway and consistent with this, a much greater proportion of cystatin F is retained in cells compared with other ‘secreted' cystatins such as cystatin C (Nathanson et al, 2002). Its restricted expression in immune cells (Ni et al, 1998) and some tumours (Morita et al, 1999) suggests that it might have important regulatory functions, making it imperative that its physiological protease targets are identified. We show here that cathepsin C is a physiological target of cystatin F but only once cystatin F has undergone post-translational proteolytic processing. Processing confers a ‘gain of function' on cystatin F and sets it apart from other family members such as cystatin C where truncation of its N terminus dramatically reduced its inhibitory activity against cathepsins B and L (Abrahamson et al, 1991).
Cathepsin C activates several important serine protease zymogens in T cells, NK cells, neutrophils and mast cells, including the granzymes A and B, cathepsin G, elastase, proteinase 3 and mast cell chymase. Activation is achieved by removal of an ‘activation dipeptide' at the N terminus (Salvesen and Enghild, 1990). The importance of cathepsin C in immunobiology is underlined by the phenotype of cathepsin C-deficient mice, which accumulate the zymogen forms of the above-mentioned substrates and fail to activate neutrophil serine proteases (Adkison et al, 2002) and mast cell chymase (Wolters et al, 2001). Defective cytotoxic T-cell effector function has also been reported in some studies (Pham and Ley, 1999) though not in others (Sutton et al, 2007). In humans, Papillon–Lefèvre syndrome occurs as a result of mutations in the cathepsin C gene and affected individuals exhibit periodontitis, skin infections and other abnormalities. Defects in resting NK cell function in Papillon-Lefèvre patients have also been found and attributed to failure to activate granzyme B (Meade et al, 2006). While cathepsin C is clearly crucial, there is also evidence that attenuation of its activity may be beneficial. Cathepsin C-deficient mice were protected from acute inflammatory arthritis due to attenuated neutrophil recruitment in the affected joints, which in turn is due to lack of elastase and cathepsin G activity (Adkison et al, 2002) and were more resistant in an aortic inflammation model likely driven by cathepsin C-dependent neutrophil serine proteases (Pagano et al, 2007). Cathepsin C-null mice were also more resistant to sepsis due to an altered mast cell-driven inflammatory response (Mallen-St Clair et al, 2004). On the basis of these and other data, cathepsin C has been proposed as a drug target for control of inflammation. Our data suggest that cystatin F may be an important endogenous cathepsin C regulator.
Our finding that cystatin F must be processed to inhibit cathepsin C can be understood in the context of the structures of cystatin–C1 cysteine protease complexes. All members of the C1 cysteine protease family have a common fold which, in the case of exopeptidases, is supplemented by additional domains that block parts of the active site. In the case of cathepsins C and H, access of the substrate beyond the S2 pocket is blocked by occluding domains not found in the endopeptidases, and these structural features determine the aminopeptidase activity of these proteases (Guncar et al, 1998; Turk et al, 2001). These domains also present a potential barrier to the docking of cystatins, which orient in the active site channel with their N-terminal region lying in and beyond the S2 pocket. In the cathepsin H/stefin A structure, the N-terminal region of stefin A is accommodated by adopting a hook-like structure, which partially displaces the mini-chain (Jenko et al, 2003). However, the cystatin F N terminus is 14 residues longer than that of stefin A and substitution of bulky residues at key places (e.g. Phe38 for Gly found in most other cystatins) may prevent a similar arrangement. Thus, full-length cystatin F can inhibit endopeptidases such as cathepsin L, which can presumably accommodate the N-terminal extension along the open substrate channel, but not aminopeptidases such as cathepsin C. We speculate that the positively charged terminal lysine of truncated cystatin F interacts with the chloride ion located at the bottom of the S2 pocket (Turk et al, 2001; Molgaard et al, 2007). The N terminus of truncated cystatin F (KPGF) is evidently a poor substrate of cathepsin C, but apparently, together with the two loop domains, a good inhibitor, a finding supported by the fact that substrates with a lysine in the P2 position are not only not cleaved, but in fact are themselves competitive inhibitors of cathepsin C (Tran et al, 2002).
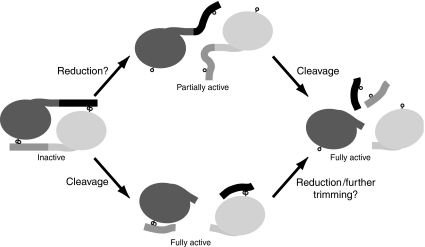
Our results suggest an alternative pathway for the activation of cystatin F in living cells involving proteolysis instead of disulphide bond reduction (Figure 7). Proteolytic cleavage at any of the residues between Cys26 and Lys35 would break the covalent linkage and hence the mutual steric hindrance between the dimer subunits. Protease binding, at least to some targets, could then occur since the short N-terminal peptide generated would be disulphide linked to Cys63 of the other subunit, well away from the protease-binding interface (Schuettelkopf et al, 2006; Figure 7). Given the high levels of reducing agent needed to activate cystatin F (Langerholc et al, 2005; Figure 1) and our finding that monomeric cystatin F had a processed N terminus (Figures 4 and 5), proteolysis might be the dominant mode of activation in cells. It will also be crucial to identify the enzyme(s) that generate the processed intracellular form of cystatin F. An intriguing possibility is that cathepsin C itself and/or its downstream protease substrates might be involved in this activation step, providing a novel mechanism to ‘self-limit' the activity of these protease systems.
Figure 7.
Pathways for cystatin F activation. Cystatin F monomers are shown in dark and light grey, with the cleaved N-terminal linking peptide in contrasting hue as shown. Cysteine residues 26 and 63 are represented by lollipops. The data favour the lower, cleavage-driven pathway of activation.
The striking coincidence of cystatin F expression in cells harbouring cathepsin C-activated granule proteases is consistent with the idea that cystatin F may attenuate this protease system. For example, in human peripheral blood, high levels of cystatin F were found in CD8+ T cells and NK cells, whereas lymphocytes whose functions do not depend on exocytosis of granule-localised effector serine proteases, such as CD4 T cells and B cells, did not express cystatin F (Figure 1). Cystatin F was also expressed in monocyte-derived dendritic cells (Figure 1; Hashimoto et al, 2000) as is cathepsin C (Hashimoto et al, 1999; Le Naour et al, 2001) To our knowledge, the function of cathepsin C in dendritic cells is not known. Conceivably, this is related to the recent demonstration that activated DCs have tumoricidal activity and express perforin and granzyme B (Stary et al, 2007).
Cystatin F colocalised with cathepsin C in murine mast cells and with the cathepsin C substrate granzyme A in human CD8 T cells. Overexpression of cystatin F in both mast cells and CD8+ T cells suppressed cathepsin C activity. However, there were clearly cystatin F-positive organelles within mast cells and CD8 T cells that lacked cathepsin C and/or its downstream substrates, and vice versa. This implies that cystatin F access to the granule system and by implication its capacity to downregulate cathepsin C activity, may be additionally regulated by inter-compartmental fusion events. In U937 cells, Langerholc et al (2005) showed cystatin F colocalises with cathepsins H and X but not with cathepsin L and other enzymes inhibited by cystatin F in vitro. Cystatin F/cathepsin colocalisation studies need to be interpreted with caution bearing in mind that cystatin F visualised by microscopy may not necessarily be in an active configuration and conversely, low amounts of active cystatin F may be difficult to visualise, but may nonetheless be functionally significant. An earlier study concluded that cystatin F did not accumulate in the endocytic pathway of U937 cells, although, consistent with our results, a dimeric form was detected in low-density Percoll gradient fractions and in the medium (Cappello et al, 2004). This apparent discrepancy can be readily explained by the fact that the antibody Cappello and colleagues used was raised against the N terminus of cystatin F, which as we show is removed by endosomal processing. Consistent with this interpretation and with our hypothesis that proteolysis is the dominant mode of activation, Cappello et al (2004) did not detect monomeric cystatin F in U937 cell lysates under non-reducing conditions.
In summary, by isolating an unusual cystatin from the specific cell types in which it is expressed, we have discovered an unexpected protease target. As an endogenous inhibitor of cathepsin C, cystatin F may attenuate the activation of a wide range of downstream serine proteases involved in inflammation and immunity. Access to the cathepsin C active site is regulated by proteolytic processing and perhaps by as yet undefined protein/membrane trafficking events. Defining how cells regulate cystatin F activation and cathepsin C interactions will be a key next step.
Materials and methods
Cell culture and isolation
Cell lines were cultured in RPMI 1640 (U937, YT, BMMC and CD8+ T cells) or DMEM (293T)-based media. DHFR-negative CHO cells were grown in IMDM-based media containing 0.1 mM hypoxanthine and 0.01 mM thymidine (HT). Following transfection with DHFR plasmids, the HT supplement was removed and methotrexate added at 0.1–10 μM, depending on the stage of selection (see Supplementary data). Mast cells and CD8+ T cells were cultured from the bone marrow and spleen, respectively, of C57Bl/6 mice. Mast cells were expanded over 4–8 weeks in media supplemented with IL-3 (10% WEHI-conditioned medium) as described (Razin et al, 1984) and purity checked by FACS analysis for Fcɛ-RI and CD117 (c-Kit) (Supplementary Figure S5). CD8+ T cells were expanded from splenocytes by stimulation with 0.5 μg/ml anti-CD3e (BD Bioscience) in the presence of 20 ng/ml recombinant human IL-2 (Chiron). Human ‘buffycoats' (Ninewells Hospital, Dundee) were used to isolate monocytic cells on Ficoll Paque (Amersham Biosciences) and B cells, CD4+ T cells, CD8+ T cells, monocytes, neutrophils and NK cells were isolated by positive selection using anti-CD19, -CD4, -CD8, -CD14, -CD16 and -CD56 magnetic beads (Miltenyi), respectively, and purity was checked by FACS staining using PE-conjugated antibodies directed against the same cell surface markers except anti-CD20 was used to assess B-cell purity. Dendritic cells were differentiated from CD14+ monocytes in GMCSF (800 U/ml; Leucomax, Sandoz) and IL-4 (1000 U/ml; BD Biosciences) essentially as described by Sallusto and Lanzavecchia (1994). DC maturation was induced with 1 μg/ml LPS and confirmed by FACS-monitored increase in CD40 and CD83.
Plasmids, transfections and retroviral transduction
Human cystatin F, additionally encoding a 6-histidine C-terminal tag, was amplified from cDNA as detailed in Supplementary data. Cystatin F with a deletion of 15 amino acids downstream of the signal sequence (Δ15N cystatin F) was generated by separate amplification of the signal sequence and residues 35–145 of cystatin F and subsequent fusion of the two PCR products. Cathepsin C and cystatin C sequences were amplified from U937 cDNA and cloned into vectors pCMV4a and pcDNA3.1, respectively. Mutagenesis of Cys26 and Cys63 of wild-type cystatin F cloned into pcDNA3.1 was performed using the QuikChange site-directed mutagenesis kit (Stratagene). Murine cystatin F was isolated from cDNA generated from CD8+ T cells and cloned into the BamHI and NotI sites of retroviral vector LZRS-pBMN (gift of Dr Gary Nolan). Full methods and all primer sequences can be found in Supplementary data.
293T cells (1 × 106 cells/10 cm dish) were transfected with 2 μg DNA using 8 μl Fugene reagent (Roche) in a final volume of 100 μl serum-free medium. DNAs for co-transfection (pCMV and pcDNA3.1 constructs) were mixed in a 1:2 ratio. Cells were analysed 48 h later. Retrovirus was produced in the packaging cell line Phoenix Eco 293T as described previously (West et al, 2004) and concentrated 10-fold. Primary spleen and bone marrow cells were transduced by ‘spinfection' (2000 r.p.m., 60 min, 25°C) and GFP-expressing cells were sorted by FACS on day 3 (CD8+ T cells) or week 4 (mast cells). Cystatin F expression was confirmed by western blot.
Cystatin F isolation and antibody production
Recombinant 6-His-tagged cystatin F or Δ15N cystatin F was expressed using a gene amplification protocol described previously (Li et al, 2003). Briefly, CHO DHFR-negative cells were transfected with the 6-His cystatin F pcDNA-DHFR plasmid and selected for growth in medium lacking HT supplement. Supernatants were initially screened by western blot using an anti-cystatin F antibody kindly provided by Dr M Abrahamson and positive clones were selected for increased cystatin F production by culture in increasing concentrations of methotrexate (0.1–10 μM) over several weeks. Cystatin F was purified from the culture medium by sequential purification on Ni-NTA agarose (Qiagen), Sephacryl S200 and Hi-Trap SP XL cation exchange resin as detailed in Supplementary data. The final yield of protein was approximately 8 mg/l of culture medium. Antibodies were raised in rabbits (Diagnostics Scotland) against either whole human or mouse cystatin F or against the N-terminal sequences GPSPDTCSQDLNSRV (human) or ARPPDFCSKDLISS (mouse) coupled via an additional C-terminal cysteine to KLH.
Isolation of cystatin F–protease complexes
Cystatin F and associated proteins were isolated from detergent lysates of U937 and YT cells using affinity-purified rabbit anti-cystatin F antibodies or control rabbit Ig coupled to Sepharose. Proteins were eluted from the washed adsorbent and separated on 4–12% Bis–Tris Novex pre-cast SDS–PAGE gels. Proteins specifically precipitated by anti-cystatin F antibodies were excised, digested with trypsin and analysed by mass spectrometry. Sequence analysis of dimeric and monomeric cellular cystatin F utilised a similar procedure except that a total membrane fraction was generated following mechanical disruption of U937 cells. Following detergent solubilisation, cystatin F was immunoprecipitated and separated by non-reducing SDS–PAGE. N-terminal sequence analysis was performed by Edman degradation. Detailed methods can be found in Supplementary data.
Protease assays
All assays were performed on a FLUOstar Optima Fluorimeter (BMG) with 360 nm excitation and 460 nm emission wavelength filters. Recombinant human cathepsins C (activated according to the manufacturer's instructions) and L were obtained from R&D Systems (Minneapolis, MN). Cathepsin C was assayed at 25°C in 25 mM MES, pH 6.0, 50 mM NaCl, 5 mM DTT, 0.1% PEG 3350 using Gly-Arg-AMC as substrate at 70 μM final concentration. Cathepsin L was assayed at 37°C in 25 mM MES, pH 6.0, 5 mM DTT, 0.1% PEG 3350 with Z-Phe-Arg-AMC as substrate (2.4 μM final concentration) in a final reaction volume of 80 μl. Reaction progress was monitored continuously by product (AMC) fluorescence and initial rates were obtained by fitting the time courses, including data with substrate depletion less than 10%. Wild-type cystatin F was pre-activated with 40 mM DTT for 1 h. Enzyme concentration was chosen to be less than the lowest inhibitor concentration used. All measurements were carried out in at least triplicate. Cathepsin activities in 10 μg of post-nuclear cell lysates were assayed using 50 μM Gly-Phe-AMC (cathepsin C) or 40 μM Z-Phe-Arg-AMC (cathepsin L) in a final volume of 200 μl 150 mM NaCl, 2 mM EDTA, 5 mM DTT, 100 mM citrate, pH 5.5. Chymase-dependent CAE activity was measured by histochemistry using naphthol AS-D CAE substrate and Fast Red Violet LB staining solution, according to the manufacturer's instructions (Sigma-Aldrich Ltd., St Louis, MO) as previously described (Wolters et al, 2001). Representative images were collected on a Zeiss LSM510 microscope using an excitation of 540 nM. The inherent fluorescent intensity of the CAE-positive cells were quantitated using Volocity software (Improvision) by dividing the sum of fluorescence intensity per field by the number of cells within that field (n>125 cells).
Metabolic labelling, subcellular fractionation and microscopy
Metabolic labelling experiments were performed essentially as described previously (Li et al, 2003). Briefly, U937 cells were preincubated in Met/Cys-free media, labelled for 20 min with [35S]Met/Cys (Amersham Biosciences) and chased in complete RPMI medium containing 100 μg/ml methionine and 500 μg/ml cysteine. Cystatin F was immunoprecipitated from aliquots of the cell lysate and medium using 2 μg of rabbit serum raised against the full-length protein. Percoll gradient fractionation of post-nuclear supernatants was performed essentially as previously reported (Davidson et al, 1990; West et al, 1994). To improve purity of lysosomal fractions (Kawashima et al, 1998), 2 mM CaCl2 was added to the post-nuclear supernatant and incubated at 37°C for 10 min prior to fractionation. Cystatin F, cathepsin C, granzyme A and markers of the endocytic and secretory pathways were localised by fluorescence microscopy in BMMC and/or CD8+ T cells as detailed in Supplementary data.
Modelling of cathepsin–cystatin complexes
Models for complexes between cystatin F and various cathepsins were constructed as described previously (Schuettelkopf et al, 2006). In brief, the structure of a cystatin F monomer (from PDB ID: 2CH9) and an appropriate cathepsin structure were superimposed on their counterparts in the crystallographic complex between stefin B and papain (PDB ID: 1STF) (Stubbs et al, 1990). The resulting crude model was manually rebuilt to remove clashes where possible and finally energy-minimised using GROMACS (Lindahl et al, 2001) with a simulated annealing protocol. For cathepsin C, PDB entry 1K3B (Turk et al, 2001) was used.
Supplementary Material
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figures S3
Supplementary Figures S4
Supplementary Figures S5
Supplementary Figure Legends
Supplementary data
Acknowledgments
This study was supported by a BBSRC CASE studentship to GH and a Wellcome Trust Programme Grant to CW. We thank David Campbell, Douglas Lamont and Kenneth Beattie for Edman sequence analysis and mass spectrometry, Rosie Clarke for FACS sorting, and John James and Alan Prescott for assistance with the microscopy. We also thank Dr Magnus Abrahamson for an initial sample of anti-cystatin F antibody.
References
- Abrahamson M, Alvarez-Fernandez M, Nathanson CM (2003) Cystatins. Biochem Soc Symp 70: 179–199 [DOI] [PubMed] [Google Scholar]
- Abrahamson M, Mason RW, Hansson H, Buttle DJ, Grubb A, Ohlsson K (1991) Human cystatin C. Role of the N-terminal segment in the inhibition of human cysteine proteinases and in its inactivation by leucocyte elastase. Biochem J 273 (Part 3): 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkison AM, Raptis SZ, Kelley DG, Pham CT (2002) Dipeptidyl peptidase I activates neutrophil-derived serine proteases and regulates the development of acute experimental arthritis. J Clin Invest 109: 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Fernandez M, Barrett AJ, Gerhartz B, Dando PM, Ni J, Abrahamson M (1999) Inhibition of mammalian legumain by some cystatins is due to a novel second reactive site. J Biol Chem 272: 19195–19203 [DOI] [PubMed] [Google Scholar]
- Bode W, Engh R, Musil D, Laber B, Stubbs M, Huber R, Turk V (1990) Mechanism of interaction of cysteine proteinases and their protein inhibitors as compared to the serine proteinase–inhibitor interaction. Biol Chem Hoppe Seyler 371 (Suppl): 111–118 [PubMed] [Google Scholar]
- Bode W, Engh R, Musil D, Thiele U, Huber R, Karshikov A, Brzin J, Kos J, Turk V (1988) The 2.0 Å X-ray crystal structure of chicken egg white cystatin and its possible mode of interaction with cysteine proteinases. EMBO J 7: 2593–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello F, Gatti E, Camossetto V, David A, Lelouard H, Pierre P (2004) Cystatin F is secreted, but artificial modification of its C-terminus can induce its endocytic targeting. Exp Cell Res 297: 607–618 [DOI] [PubMed] [Google Scholar]
- Dahl SW, Halkier T, Lauritzen C, Dolenc I, Pedersen J, Turk V, Turk B (2001) Human recombinant pro-dipeptidyl peptidase I (cathepsin C) can be activated by cathepsins L and S but not by autocatalytic processing. Biochemistry 40: 1671–1678 [DOI] [PubMed] [Google Scholar]
- Davidson HW, West MA, Watts C (1990) Endocytosis, intracellular trafficking and processing of membrane IgG and monovalent antigen/membrane IgG complexes in B lymphocytes. J Immunol 144: 4101–4109 [PubMed] [Google Scholar]
- Guncar G, Podobnik M, Pungercar J, Strukelj B, Turk V, Turk D (1998) Crystal structure of porcine cathepsin H determined at 2.1 Å resolution: location of the mini-chain C-terminal carboxyl group defines cathepsin H aminopeptidase function. Structure 6: 51–61 [DOI] [PubMed] [Google Scholar]
- Halfon S, Ford J, Foster J, Dowling L, Lucian L, Sterling M, Xu Y, Weiss M, Ikeda M, Liggett D, Helms A, Caux C, Lebecque S, Hannum C, Menon S, McClanahan T, Gorman D, Zurawski G (1998) Leukocystatin, a new class II cystatin expressed selectively by hematopoietic cells. J Biol Chem 273: 16400–16408 [DOI] [PubMed] [Google Scholar]
- Hashimoto S, Suzuki T, Dong HY, Nagai S, Yamazaki N, Matsushima K (1999) Serial analysis of gene expression in human monocyte-derived dendritic cells. Blood 94: 845–852 [PubMed] [Google Scholar]
- Hashimoto SI, Suzuki T, Nagai S, Yamashita T, Toyoda N, Matsushima K (2000) Identification of genes specifically expressed in human activated and mature dendritic cells through serial analysis of gene expression. Blood 96: 2206–2214 [PubMed] [Google Scholar]
- Jedeszko C, Sloane BF (2004) Cysteine cathepsins in human cancer. Biol Chem 385: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Jenko S, Dolenc I, Guncar G, Dobersek A, Podobnik M, Turk D (2003) Crystal structure of Stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases. J Mol Biol 326: 875–885 [DOI] [PubMed] [Google Scholar]
- Kawashima A, Sato A, Kawashima M, Nitta K, Yumuran W, Sugino N, Nihei H, Natori Y (1998) A simple procedure for the isolation of rat kidney lysosomes. Kidney Int 54: 275–278 [DOI] [PubMed] [Google Scholar]
- Kopitar-Jerala N (2006) The role of cystatins in cells of the immune system. FEBS Lett 580: 6295–6301 [DOI] [PubMed] [Google Scholar]
- Langerholc T, Zavasnik-Bergant V, Turk B, Turk V, Abrahamson M, Kos J (2005) Inhibitory properties of cystatin F and its localization in U937 promonocyte cells. FEBS J 272: 1535–1545 [DOI] [PubMed] [Google Scholar]
- Le Naour F, Hohenkirk L, Grolleau A, Misek DE, Lescure P, Geiger JD, Hanash S, Beretta L (2001) Profiling changes in gene expression during differentiation and maturation of monocyte-derived dendritic cells using both oligonucleotide microarrays and proteomics. J Biol Chem 276: 17920–17931 [DOI] [PubMed] [Google Scholar]
- Li DN, Matthews SP, Antoniou AN, Mazzeo D, Watts C (2003) Multistep autoactivation of asparaginyl endopeptidase in vitro and in vivo. J Biol Chem 278: 38980–38990 [DOI] [PubMed] [Google Scholar]
- Lindahl E, Hess B, van der Spoel D (2001) Gromacs 3.0: a package for molecular simulation and trajectory analysis. J Mol Mod 7: 306–317 [Google Scholar]
- Lindahl P, Nycander M, Ylinenjarvi K, Pol E, Bjork I (1992) Characterization by rapid-kinetic and equilibrium methods of the interaction between N-terminally truncated forms of chicken cystatin and the cysteine proteinases papain and actinidin. Biochem J 286 (Part 1): 165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sukhova GK, Sun JS, Xu WH, Libby P, Shi GP (2004) Lysosomal cysteine proteases in atherosclerosis. Arterioscler Thromb Vasc Biol 24: 1359–1366 [DOI] [PubMed] [Google Scholar]
- Mallen-St Clair J, Pham CT, Villalta SA, Caughey GH, Wolters PJ (2004) Mast cell dipeptidyl peptidase I mediates survival from sepsis. J Clin Invest 113: 628–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallen-St Clair J, Shi GP, Sutherland RE, Chapman HA, Caughey GH, Wolters PJ (2006) Cathepsins L and S are not required for activation of dipeptidyl peptidase I (cathepsin C) in mice. Biol Chem 387: 1143–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoury B, Gregory WF, Maizels RM, Watts C (2001) Bm-CPI-2, a cystatin homolog secreted by the filarial parasite Brugia malayi, inhibits class II MHC-restricted antigen processing. Curr Biol 11: 447–451 [DOI] [PubMed] [Google Scholar]
- Meade JL, de Wynter EA, Brett P, Sharif SM, Woods CG, Markham AF, Cook GP (2006) A family with Papillon–Lefevre syndrome reveals a requirement for cathepsin C in granzyme B activation and NK cell cytolytic activity. Blood 107: 3665–3668 [DOI] [PubMed] [Google Scholar]
- Molgaard A, Arnau J, Lauritzen C, Larsen S, Petersen G, Pedersen J (2007) The crystal structure of human dipeptidyl peptidase I (cathepsin C) in complex with the inhibitor Gly-Phe-CHN2. Biochem J 401: 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Yoshiuchi N, Arakawa H, Nishimura S (1999) CMAP: a novel cystatin-like gene involved in liver metastasis. Cancer Res 59: 151–158 [PubMed] [Google Scholar]
- Nathanson CM, Wasselius J, Wallin H, Abrahamson M (2002) Regulated expression and intracellular localization of cystatin F in human U937 cells. Eur J Biochem 269: 5502–5511 [DOI] [PubMed] [Google Scholar]
- Ni J, Fernandez A, Danielsson L, Chillakuru RA, Zhang J, Grub A, Su J, Gentz R, Abrahamson M (1998) Cystatin F is a glycosylated human low molecular weight cystein proteinase inhibitor. J Biol Chem 273: 24797–24804 [DOI] [PubMed] [Google Scholar]
- Obata-Onai A, Hashimoto S, Onai N, Kurachi M, Nagai S, Shizuno K, Nagahata T, Matsushima K (2002) Comprehensive gene expression analysis of human NK cells and CD8(+) T lymphocytes. Int Immunol 14: 1085–1098 [DOI] [PubMed] [Google Scholar]
- Pagano MB, Bartoli MA, Ennis TL, Mao D, Simmons PM, Thompson RW, Pham CT (2007) Critical role of dipeptidyl peptidase I in neutrophil recruitment during the development of experimental abdominal aortic aneurysms. Proc Natl Acad Sci USA 104: 2855–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham CT, Ley TJ (1999) Dipeptidyl peptidase I is required for the processing and activation of granzymes A and B in vivo. Proc Natl Acad Sci USA 96: 8627–8632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin E, Ihle JN, Seldin D, Mencia-Huerta JM, Katz HR, LeBlanc PA, Hein A, Caulfield JP, Austen KF, Stevens RL (1984) Interleukin 3: a differentiation and growth factor for the mouse mast cell that contains chondroitin sulfate E proteoglycan. J Immunol 132: 1479–1486 [PubMed] [Google Scholar]
- Sallusto F, Lanzavecchia A (1994) Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med 179: 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen G, Enghild JJ (1990) An unusual specificity in the activation of neutrophil serine proteinase zymogens. Biochemistry 29: 5304–5308 [DOI] [PubMed] [Google Scholar]
- Schuettelkopf AW, Hamilton G, Watts C, van Aalten DM (2006) Structural basis of reduction-dependent activation of human cystatin F. J Biol Chem 281: 16570–16575 [DOI] [PubMed] [Google Scholar]
- Stary G, Bangert C, Tauber M, Strohal R, Kopp T, Stingl G (2007) Tumoricidal activity of TLR7/8 activated inflammatory dendritic cells. J Exp Med 204: 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs MT, Laber B, Bode W, Huber R, Jerala R, Lenarcic B, Turk V (1990) The refined 2.4 Å X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J 9: 1939–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton VR, Waterhouse NJ, Browne KA, Sedelies K, Ciccone A, Anthony D, Koskinen A, Mullbacher A, Trapani JA (2007) Residual active granzyme B in cathepsin C-null lymphocytes is sufficient for perforin-dependent target cell apoptosis. J Cell Biol 176: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TV, Ellis KA, Kam CM, Hudig D, Powers JC (2002) Dipeptidyl peptidase I: importance of progranzyme activation sequences, other dipeptide sequences, and the N-terminal amino group of synthetic substrates for enzyme activity. Arch Biochem Biophys 403: 160–170 [DOI] [PubMed] [Google Scholar]
- Turk D, Janjic V, Stern I, Podobnik M, Lamba D, Dahl SW, Lauritzen C, Pedersen J, Turk V, Turk B (2001) Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J 20: 6570–6582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MA, Lucocq JM, Watts C (1994) Antigen processing and class II MHC peptide-loading compartments in human B-lymphoblastoid cells [see comments]. Nature 369: 147–151 [DOI] [PubMed] [Google Scholar]
- West MA, Wallin RP, Matthews SP, Svensson HG, Zaru R, Ljunggren HG, Prescott AR, Watts C (2004) Enhanced dendritic cell antigen capture via toll-like receptor-induced actin remodeling. Science 305: 1153–1157 [DOI] [PubMed] [Google Scholar]
- Wolters PJ, Pham CT, Muilenburg DJ, Ley TJ, Caughey GH (2001) Dipeptidyl peptidase I is essential for activation of mast cell chymases, but not tryptases, in mice. J Biol Chem 276: 18551–18556 [DOI] [PubMed] [Google Scholar]
- Zeeuwen PL, van Vlijmen-Willems IM, Hendriks W, Merkx GF, Schalkwijk J (2002) A null mutation in the cystatin M/E gene of ichq mice causes juvenile lethality and defects in epidermal cornification. Hum Mol Genet 11: 2867–2875 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figures S3
Supplementary Figures S4
Supplementary Figures S5
Supplementary Figure Legends
Supplementary data