Abstract
MgtC is a virulence factor common to several intracellular pathogens that is required for intramacrophage survival and growth in magnesium-depleted medium. In Salmonella enterica, MgtC is coexpressed with the MgtB magnesium transporter and transcription of the mgtCB operon is induced by magnesium deprivation. Despite the high level of mgtCB transcriptional induction in magnesium-depleted medium, the MgtC protein is hardly detected in a wild-type Salmonella strain. Here, we show that downregulation of MgtC expression is dependent on a hydrophobic peptide, MgtR, which is encoded by the mgtCB operon. Our results suggest that MgtR promotes MgtC degradation by the FtsH protease, providing a negative regulatory feedback. Bacterial two-hybrid assays demonstrate that MgtR interacts with the inner-membrane MgtC protein. We identified mutant derivatives of MgtR and MgtC that prevent both regulation and interaction between the two partners. In macrophages, overexpression of the MgtR peptide led to a decrease of the replication rate of Salmonella. This study highlights the role of peptides in bacterial regulatory mechanisms and provides a natural antagonist of the MgtC virulence factor.
Keywords: FtsH, MgtC, regulatory peptide, Salmonella enterica serovar Typhimurium
Introduction
Bacteria have evolved sophisticated regulatory pathways to adapt to environmental changes. The control of protein stability allows the cell to adjust to changing conditions by changing the rate of turnover of proteins. Regulated proteolysis has been well characterized for few proteins, including proteins involved in stress response mechanisms (Gottesman, 2003). Whereas the complex ubiquitin ligase machinery provides a target for regulating proteolysis in eukaryotes, no ubiquitin tagging exists in bacteria. A small number of ATP-dependent proteases (AAA+ proteases) are responsible for degradation of specific proteins, frequently by directly interacting with the substrate protein (Baker and Sauer, 2006). An additional layer of potential specificity and regulation is provided by adaptor proteins, that enhance or alter the substrate-recognition properties of AAA+ proteases (Dougan et al, 2002; Baker and Sauer, 2006). The ClpXP and Lon AAA+ proteases have been implicated in bacterial virulence (Yamamoto et al, 2001; Frees et al, 2003; Takaya et al, 2003; Kwon et al, 2004) and in the degradation of transcriptional regulators that control expression of virulence gene (Takaya et al, 2005).
MgtC is a virulence factor common to several intracellular pathogens that plays a key role in intramacrophage survival (Alix and Blanc-Potard, 2007). MgtC was first described in Salmonella enterica serovar Typhimurium (S. typhimurium) where it is required for intramacrophage multiplication and long-term systemic infection in mice (Blanc-Potard and Groisman, 1997; Lawley et al, 2006). MgtC is also a critical factor for the intramacrophage growth of Mycobacterium tuberculosis, Brucella suis, Yersinia pestis and Burkholderia cenocepacia (Buchmeier et al, 2000; Lavigne et al, 2005; Grabenstein et al, 2006; Maloney and Valvano, 2006). Phylogenetic analysis suggested that mgtC has been acquired by horizontal gene transfer repeatedly throughout bacterial evolution (Blanc-Potard and Lafay, 2003). In S. typhimurium, the mgtC gene is carried by the SPI-3 pathogenicity island (Blanc-Potard and Groisman, 1997). The biochemical function of MgtC remains unknown and the role of MgtC might differ in host and non-host environments (Rang et al, 2007). Heterologous expression of MgtC protein in Xenopus laevis oocytes has significant effect on cellular ion homeostasis via a modulation of eukaryotic Na+,K+-ATPase activity (Günzel et al, 2006). However, the physiological relevance of these results during bacterial infection is currently unknown.
MgtC is also involved in adaptation to low-Mg2+ environments (Alix and Blanc-Potard, 2007). The regulation of mgtC in response to Mg2+ deprivation has been well studied at the transcriptional level in S. typhimurium. The S. typhimurium mgtC gene is co-transcribed with mgtB, encoding an Mg2+ transporter (Snavely et al, 1991). Both MgtC and MgtB are inner-membrane proteins. The mgtCB operon is regulated by the PhoP–PhoQ two-component system and is highly induced by low Mg2+ concentration (García-Véscovi et al, 1996). In addition, the mgtCB operon is also regulated at the transcriptional level by a PhoPQ-independent mechanism that depends on the secondary structure of the 5′UTR (Groisman et al, 2006). The MgtC protein could be detected at short time periods after Mg2+ starvation (Moncrief and Maguire, 1998; Adkins et al, 2006). Surprisingly, despite the high transcriptional induction, MgtC is undetectable during prolonged Mg2+ starvation in an mgtC+ mgtB+ strain, although large amounts of MgtB are observed (Moncrief and Maguire, 1998). However, MgtC was detected in an mgtC+ strain that lacks the mgtB sequence, suggesting that MgtC is not produced or is not stable in the presence of the MgtB protein (Moncrief and Maguire, 1998). In the present study, we further investigated the regulation of MgtC expression. We demonstrate that the amount of MgtC protein is downregulated by a 30-amino-acid peptide that is encoded downstream of the mgtB gene. We show that this peptide, termed MgtR, interacts with MgtC in vivo and promotes the degradation of MgtC by the FtsH AAA+ protease. Identification of MgtR highlights the regulatory role of peptides and very small proteins that are often missed in genomic and proteomic analyses.
Results
MgtC expression is negatively regulated by sequences 3′ of mgtB
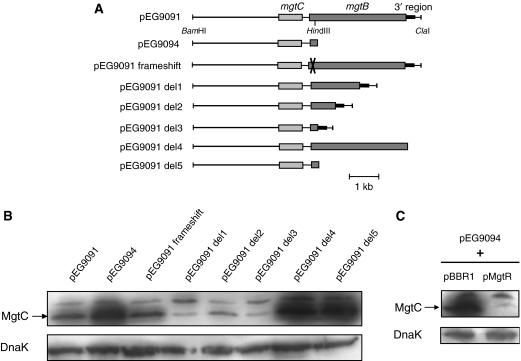
Previous work has shown that even though the mgtCB operon is highly induced in low-Mg2+ environment, the MgtB, but not the MgtC, protein is detected in a wild-type Salmonella strain upon prolonged Mg2+ starvation (Moncrief and Maguire, 1998). In addition, it has been suggested that mgtB sequence has a negative effect on MgtC expression (Moncrief and Maguire, 1998). To explain the role of mgtB on MgtC protein level, we have transformed an Escherichia coli strain, which does not contain the mgtCB operon, with plasmids containing only the mgtC gene (pEG9094) or the entire mgtCB operon (pEG9091) (Figure 1A). Consistent with previous data, MgtC was clearly expressed from pEG9094, but to a much lesser extent from pEG9091 (Figure 1B). A frameshift mutation at the beginning of the mgtB sequence did not produce higher MgtC levels, indicating that the lack of MgtC expression from pEG9091 is not due to the production of MgtB protein per se, but rather to other elements in the mgtB sequence (Figure 1B). This result agrees with the previous report that a functional MgtB protein was not required to repress MgtC expression (Moncrief and Maguire, 1998). We carried out successive deletions on pEG9091 plasmid to characterize the DNA region responsible for down-regulation of MgtC expression (Figure 1A). Deletions within the mgtB gene did not increase MgtC levels (Figure 1B). However, a deletion of the region downstream of the mgtB coding region (pEG9091 del4) gave a pattern similar to the one observed with pEG9094, indicating that the 3′ region of mgtB is involved in the regulation of MgtC expression.
Figure 1.
The levels of the MgtC protein are regulated by the 3′ region of the mgtCB operon. (A) Schematic representation of sequences carried by plasmids pEG9091 (mgtC+ mgtB+), pEG9094 (mgtC+) and pEG9091 derivatives. (B) Western blot analysis of E. coli strains transformed with pEG9091, pEG9094 or pEG9091 derivatives. Total extracts were prepared from bacteria grown for 16 h in low-Mg2+ medium and were blotted with anti-MgtC antibodies or anti-DnaK antibodies. The band detected above MgtC appears non-specific since it is also found with E. coli strains or S. typhimurium strain NM14. (C) Western blot analysis of E. coli strains transformed with pEG9094 and pBBR1MCS vector or pMgtR. The pMgtR plasmid is a pBBR1MCS derivative that carries a sequence of 150 bp downstream of mgtB (which is highlighted in black in panel A).
A 150-bp region downstream of mgtB was subcloned into the pBBR1MCS vector plasmid, which is compatible with pEG9094, to produce the pMgtR plasmid. The pMgtR plasmid was introduced in E. coli strain carrying pEG9094. Western blot analysis clearly showed a decrease of MgtC expression in the presence of pMgtR but not the pBBR1MCS vector (Figure 1C). This result indicates that the region downstream of mgtB acts in trans to regulate MgtC expression. Taken together, these results indicate that a small DNA region, present at the 3′ end of the mgtCB operon, negatively regulates the amount of MgtC protein.
MgtR: a peptide that downregulates MgtC expression
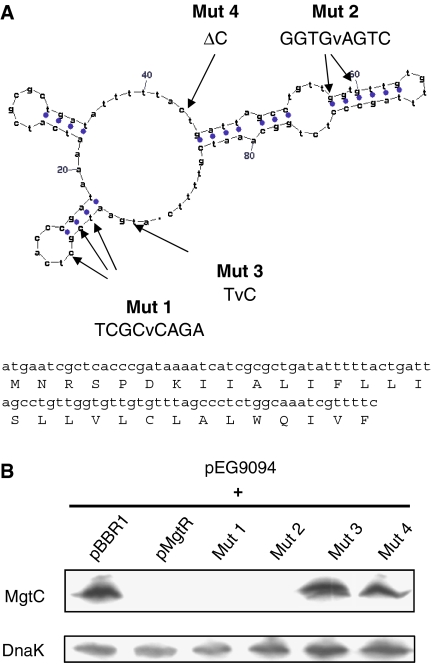
The 150-bp regulatory region described above includes a 90-bp region (position 3961428–3961518 of the S. typhimurium LT2 genome) that could encode a small RNA and/or an open reading frame (ORF) of 30 amino acids, preceded by a shine–dalgarno consensus sequence (Figure 2A). This sequence, which is not annotated in the S. typhimurium genome, was named mgtR (GenBank accession number EU154350). To distinguish between the RNA and protein hypotheses, we examined the phenotype of strains with several site-directed mutants in the pMgtR plasmid. First, qwe generated two mutations (Mut 1 and Mut 2) that disrupt the predicted RNA secondary structure without changing the predicted amino-acid sequence (Figure 2A). As shown in Figure 2B, regulation of MgtC expression was maintained in the presence of both mutations. Secondly, we performed mutagenesis of the initial ATG of the putative ORF (Mut 3) or introduced a frameshift mutation (Mut 4), without changing the stability of the predicted RNA secondary structure (Figure 2A). The amount of MgtC was highly increased when pMgtR plasmid carried these mutations (Figure 2B), indicating that these mutations prevented the down-regulation of MgtC expression. Taken together, these results indicate that a peptide, and not a small RNA, is involved in MgtC regulation.
Figure 2.
The 3′ region of the mgtCB operon encodes a peptide that is involved in MgtC downregulation. (A) The 150-bp regulatory region at the 3′ end of the mgtCB operon includes a 90-bp sequence that could encode a small RNA or a peptide. One of the secondary structures of a putative RNA predicted by Mfold server (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html) is shown (dG=−15.5 kcal mole−1). Site-directed mutagenesis was performed on pMgtR plasmid at positions 1 (TCGCvCAGA) and 2 (GGTGvAGTC) to destabilize RNA structure without affecting protein sequence. The mutations 3 (TvC) and 4 (ΔC) affected synthesis of a putative ORF, without affecting predicted RNA structure. (B) Western blot analysis of E. coli strains transformed with pEG9094 and mutated pMgtR plasmids. The pBBR1MCS vector and pMgtR plasmid were used as controls. Total extracts were prepared from bacteria grown for 16 h in low-Mg2+ medium and were blotted with anti-MgtC antibodies or anti-DnaK antibodies.
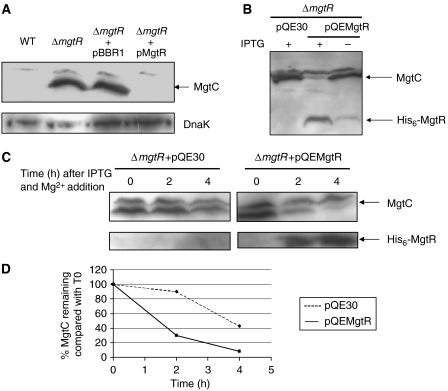
To further explore the role of mgtR in S. typhimurium, we have carried out a precise chromosomal deletion of mgtR. As expected, MgtC is expressed in the ΔmgtR-mutant strain and expression is turned down by introduction of the pMgtR plasmid (Figure 3A). The mgtR gene was cloned into pQE30 to produce an IPTG-inducible N-terminal His-tagged peptide. The resulting plasmid, pQEMgtR, produces a functional peptide since it complemented the S. typhimurium ΔmgtR-mutant strain upon IPTG induction by downregulating MgtC expression (Figure 3B). Upon IPTG induction, the decrease in the level of MgtC was correlated with the detection of the 5-kDa His6-MgtR at the bottom of the gel. The detection of His6-MgtR in this western blot experiment is due to the fact that MgtC antibodies recognize the His epitope. The 5-kDa band is also detected upon IPTG induction in a strain that encodes His6-MgtR but not MgtC, indicating that it is not related with degradation fragments of MgtC (data not shown). Taken together, these results demonstrate that a 30-amino-acid peptide encoded downstream of mgtB negatively regulates the levels of MgtC protein.
Figure 3.
Downregulation of MgtC by the induction of a His6-MgtR peptide in a Salmonella ΔmgtR mutant. Western blot experiments were carried out on Salmonella extracts using anti-MgtC antibodies. (A) Characterization of a Salmonella ΔmgtR mutant and complementation with the pMgtR plasmid. Western blot on S. typhimurium wild-type and ΔmgtR-mutant strains. The pMgtR plasmid or the pBBR1MCS vector was introduced into the ΔmgtR-mutant strain. Extracts were blotted with anti-DnaK antibodies as control. (B) Induction of His6-MgtR expression by addition of 0.1 mM IPTG decreased the expression of MgtC in a Salmonella ΔmgtR-mutant strain. The lower band is the His6-MgtR peptide (approximately 5 kDa), which cross-reacts with anti-MgtC antibodies. (C) Kinetic of MgtC degradation by His6-MgtR. After overnight growth without IPTG in conditions of mgtC transcription (10 μM Mg2+), bacteria were shifted to a medium containing 10 mM Mg2+ to shut down mgtC transcription and IPTG to induce His6-MgtR expression. In each case, samples were prepared for Western blot at several time periods after the shift (0, 2 and 4 h). Identical volumes corresponding to a constant number of cells were loaded independently of the time point. (D) Quantification of the levels of MgtC in panel C.
The mgtR sequence belongs to the mgtCB operon and MgtR acts on MgtC at the post-transcriptional level
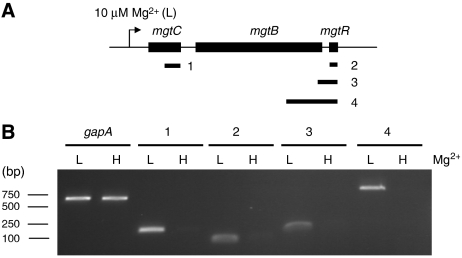
The mgtCB operon is induced by growth in low-Mg2+ medium (Figure 4A). To examine mgtR transcription, total RNA was extracted from wild-type S. typhimurium strain 14028s grown in NCE medium supplemented with 10 μM or 10 mM Mg2+. RT–PCR experiments showed that, similarly to mgtC, the mgtR gene is only transcribed in low-Mg2+ conditions. In contrast, a non-PhoP-regulated gene, gapA, is transcribed in both low and high Mg2+ (Figure 4B). In addition, RT–PCR fragments were amplified with primers belonging to mgtB and mgtR (Figure 4B). Taken together, these results demonstrate that mgtR belongs to the mgtCB operon.
Figure 4.
The mgtR sequence belongs to mgtCB operon. RT–PCR experiments were performed on total RNA of wild-type S. typhimurium extracted after 30-min growth with 10 μM Mg2+ (L) or with 10 mM Mg2+ (H). (A) Schematic representation of the fragments amplified by RT–PCR. (B) Analysis of DNA fragments amplified by RT–PCR on agarose gel. RT–PCR of gapA is used as control. No band was amplified in the absence of reverse transcriptase (not shown).
The levels of MgtB protein were identical in the presence or absence of MgtR (Supplementary Figure S1), indicating that the effect of MgtR is specific for MgtC and does not affect MgtB. This result further suggests that MgtR does not act on the promoter sequences of the mgtCB operon. Accordingly, when the region upstream of mgtC, including mgtC promoter sequence and mgtC 5′UTR, was fused to the lacZ gene (Supplementary data), the β-galactosidase activity was independent of the presence of MgtR (data not shown). Moreover, MgtC expression was regulated by MgtR when the mgtC coding sequence was under the control of the lac promoter (data not shown). Cumulatively, these results indicate that MgtR acts on MgtC at the post-transcriptional level.
MgtR promotes MgtC degradation by FtsH protease
We have studied the stability of MgtC in an S. typhimurium ΔmgtR-mutant strain that harbours the inducible His6-MgtR peptide. Bacterial cultures were grown O/N in low-Mg2+ medium to induce MgtC expression. The transcription of mgtC was then stopped by shifting the bacteria to high-Mg2+ medium. In addition, IPTG was simultaneously added to induce the His6-MgtR synthesis. In a control experiment, the ΔmgtR strain harboured the pQE30 vector and did not express MgtR. The level of MgtC protein was evaluated at time points after the shift (Figure 3C). In the presence of MgtR, MgtC was barely detected 4 h after the shift. On the other hand, MgtC was still detected 4 h after the shift in the absence of MgtR. This result demonstrates that MgtR promotes MgtC degradation, although through a relatively slow process.
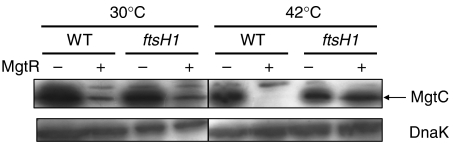
FtsH is a AAA+ protease that has been involved in the degradation of membrane proteins (Ito and Akiyama, 2005). To determine whether FtsH plays a role in MgtC degradation, we have studied the level of MgtC protein in the presence or absence of MgtR in an E. coli thermosensitive ftsH mutant or an isogenic wild-type strain. At the permissive temperature of 30°C, the down-regulation of MgtC in the presence of MgtR is similar in the wild-type and ftsH mutant (Figure 5). However, at 42°C, the regulation is clearly observed in the wild-type strain but not in the ftsH mutant (Figure 5). Hence, a functional FtsH protease is required to mediate the downregulation of MgtC expression in the presence of MgtR. To address the specificity of the FtsH protease, we have also used a clpX mutant of Salmonella (Aldridge et al, 2003). The regulation of MgtC by MgtR was similar in the wild-type strain and the clpX1∷Tn10dcam mutant (Supplementary Figure S2). Taken together, these results indicate that MgtR assists MgtC to be degraded by the FtsH protease.
Figure 5.
The FtsH protease is involved in MgtC negative regulation mediated by MgtR. The thermosensitive E. coli ftsH mutant and the isogenic ftsH+ strain were transformed by plasmids pEG9094 (mgtC+) or pEG9091 del3 (mgtC+ mgtR+) (Figure 1A). Strains were cultured in low-Mg2+ medium for 36 h at 30°C or for 7 h at 30°C before an 18-h shift at 42°C. Total extracts were blotted with anti-MgtC antibodies or anti-DnaK antibodies.
Identification of MgtR and MgtC mutants that prevent MgtR-assisted MgtC degradation
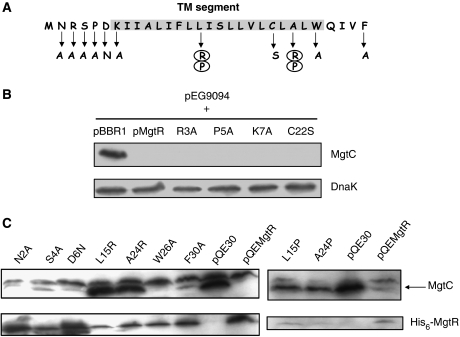
MgtR is predicted to be an inner-membrane peptide of 3.5 kDa with a short cytoplasmic N-terminus, a highly hydrophobic transmembrane α-helix and a short periplasmic C-terminus. As expected, MgtR is found to be associated with the S. typhimurium membrane fraction (Supplementary Figure S3). We carried out site-directed mutagenesis in the N- and C-terminal parts as well as in the transmembrane helix (Figure 6A). We tested the ability of mutated peptides to regulate MgtC degradation. Mutations were carried out on the pMgtR plasmid (Figure 6B) or on the pQEMgtR plasmid (Figure 6C). Results clearly showed that, among the 11 residues tested, only two residues abolished the degradation of MgtC and are therefore important for MgtR function (Figure 6C). These mutations, L15R and A24R, introduce charged residues in the hydrophobic region. On the other hand, single mutations in the N-terminal or C-terminal extremities had minor or no detectable effect on MgtR function. In the L15R mutant of MgtR, the peptide was poorly expressed and was similarly found in insoluble and soluble fractions, suggesting incorrect MgtR location (Figure 6C; Supplementary Figure S3). The A24R mutant of MgtR appeared mostly to be membrane associated despite the introduction of a charged residue. We have carried out additional mutations, A15P and A24P, that would disrupt the α-helix but maintain the hydrophobicity. Both mutations reduced MgtC degradation (Figure 6C).
Figure 6.
Amino-acid residues of MgtR important for MgtC degradation. (A) Position of the mutations on MgtR sequence. The predicted transmembrane domain is shaded. Changes that lead to loss of function are highlighted with circles. (B) Western blot analysis of crude extracts prepared from strains with mutant MgtR encoded by pMgtR derivatives. Extracts were prepared from E. coli strains carrying the pEG9094 plasmid (mgtC+) and the mutated pMgtR plasmids. The pBBR1MCS vector and pMgtR plasmid were used as control. Total extracts were blotted with anti-MgtC antibodies or anti-DnaK antibodies. (C) Western blot analysis of crude extracts prepared from strains with mutant MgtR encoded by pQEMgtR derivatives. Extracts were prepared from S. typhimurium ΔmgtR strain carrying the mutated pQEMgtR plasmids. The pQE30 vector and pQEMgtR plasmid were used as control. Total extracts were blotted with anti-MgtC antibodies that recognize both MgtC and His6-MgtR.
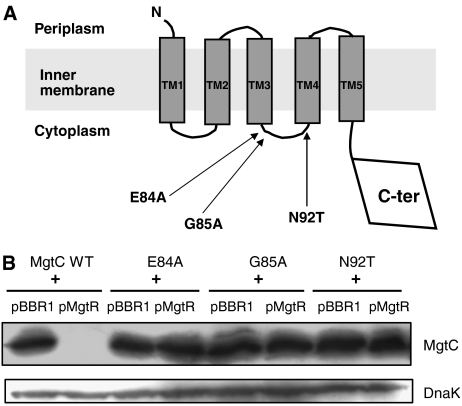
In the course of previous studies, we have characterized several mutant forms of MgtC substituted in residues that are conserved among MgtC proteins (Rang et al, 2007; C Rang and A-B Blanc-Potard, unpublished results). We have noticed that mutants E84A, G85A and N92T, led to a high level of MgtC protein by western blot analysis in S. typhimurium (data not shown). We hypothesized that these mutations, which are clustered in the cytoplasmic loop between the third and fourth transmembrane domains (TM3 and TM4) of MgtC (Figure 7A), might interfere with MgtR regulation. Plasmids encoding mutant MgtC proteins were introduced in isogenic E. coli strains expressing or lacking MgtR. In contrast to the wild-type protein, mutant MgtC proteins were similarly expressed in the presence or absence of MgtR (Figure 7B). Hence, the E84A, G85A and N92T mutations render MgtC resistant to degradation promoted by MgtR.
Figure 7.
Identification of MgtC mutants that are insensitive to downregulation by MgtR. (A) Schematic representation of MgtC protein topology. Mutations that promoted MgtC expression on Western blot are clustered in the second cytoplasmic loop. (B) Western blot analysis of MgtC mutants. MgtC mutations were carried out on the pNM12 plasmid. Extracts were prepared from E. coli strains carrying pMgtR plasmid or the pBBR1MCS vector, and the pNM12 plasmid (mgtC+) or the mutated pNM12 derivatives.
Bacterial two-hybrid analysis demonstrates that MgtC and MgtR interact in vivo
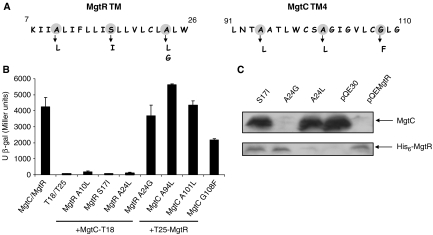
Both MgtC and MgtR are predicted to localize in the inner-membrane. To test putative interaction between MgtC and MgtR, we have used the Bacterial Adenylate Cyclase Two-Hybrid (BACTH) system that has been validated for detecting specific interactions between inner-membrane proteins (Karimova et al, 2005). A derivative of pUT18 was constructed to fuse the T18 fragment of adenylate cyclase to the C-terminal end of MgtC, that is cytoplasmic (Rang et al, 2007). A derivative of pKT25ΔXbaI was constructed to fuse the T25 fragment of adenylate cyclase to the N-terminal end of MgtR, that is predicted to be cytoplasmic. Plamids encoding the T18 and T25 fusion proteins were introduced in an E. coli cya mutant (BTH101) and functional complementation was determined by measuring β-galactosidase activity (Figure 8). A high level of β-galactosidase activity was observed when BTH101 was co-transformed with the MgtC-T18 and T25-MgtR encoding plasmids, indicating an interaction between both fusion proteins. Only basal level of β-galactosidase activity was observed with the two vectors or with either fusion protein with the other vector control.
Figure 8.
Analysis of in vivo interaction between MgtC and MgtR using the BACHT system. The E. coli BTH101 strain was co-transformed with plasmids encoding MgtC-T18 and T25-MgtR fusions. Assays were carried out at 30°C. Four mutations, L15R, L15P, A24R and A24P, were introduced in the T25-MgtR fusion. Three mutations, E84A, G85A and N92T were introduced in the MgtC-T18 fusion. The basal level of β-galactosidase activity measured with vector is approximately 60 Miller units.
To investigate the correlation between MgtC–MgtR interaction and MgtC downregulation, we have introduced in the two-hybrid plasmids mutations in MgtR or MgtC that prevent MgtR-assisted MgtC degradation. As shown in Figure 8, the two mutations in the hybrophobic domain of MgtR, L15R and A24R gave only a basal level of β-galactosidase activity, indicating a lack of MgtR–MgtC interaction, whereas mutations L15P and A24P only lowered the interaction. The substitutions E84A, G85A and N92T in MgtC also lowered the interaction, with a marked effect for the MgtC mutant G85A that exhibited a β-galactosidase activity only 2.5-fold higher than basal level. These effects are not due to a lower expression of the mutated hybrid proteins (data not shown). The effect on the interaction of mutations L15P and A24P in MgtR and E84A and N92T in MgtC appeared much clearer when the β-galactosidase assays were carried out at a more stringent temperature, 37°C, since the level of β-galactosidase was below the threshold for interaction (Supplementary Figure S4). Taken together, these results show that MgtC and MgtR mutants that prevent the degradation of MgtC can be correlated with a decrease of in vivo interaction between the two partners.
The fact that MgtC residues located in the cytoplasmic loop between TM3 and TM4 play a role in the interaction with MgtR suggests that TM3 and/or TM4 might interact with MgtR. Both MgtR and TM4 contain cysteine residues. We have shown that the C22 residue of MgtR had no role in regulation (Figure 6B) and a C99A mutation in the TM4 of MgtC had no effect on regulation and interaction (data not shown). MgtR and the TM4 domain of MgtC also harbour an Ala-coil domain, which is an helix-helix interaction motif characterized by small residues (Ala, Gly, Ser) in heptad repeats (Walters and DeGrado, 2006). To test the contribution of Ala-coil motifs in MgtC–MgtR interaction, we have substituted small residues of the motif by large hydrophobic residues (Figure 9A). Results clearly showed that the disruption of MgtR Ala-coil prevented the interaction (Figure 9B). The reduced interaction is not linked to inaccurate expression or location of MgtR since, for example, an MgtR peptide carrying mutation S17I is produced and localized like wild-type MgtR (Supplementary Figure S3). Conversely, the change to another small residue (mutation A24G) had no effect (Figure 9B), probably because a correct helix packing is preserved. A correlation is found between MgtC–MgtR interaction with the BACTH system and regulation of MgtC expression in western blot analysis (Figure 9C). On the other hand, single mutations in the Ala-coil domain of MgtC TM4 had no effect on MgtC–MgtR interaction with the exception of G108F (Figure 9B; Supplementary Figure S4). The loose interaction in the presence of G108F mutation might be related to a lower expression of the MgtC-T18 hybrid protein (data not shown).
Figure 9.
Role of Ala-coil motifs in MgtC–MgtR interaction. (A) Ala-coil motifs and mutagenesis on MgtR and MgtC TM4 sequences. (B) Effect of Ala-coil mutations on the interaction between MgtC and MgtR in vivo using the BACHT system. Assays were carried out at 30°C. (C) Effect of Ala-coil mutations in MgtR on MgtC expression. Extracts were prepared from S. typhimurium ΔmgtR strain carrying the mutated pQEMgtR plasmids and blotted with anti-MgtC antibodies. The pQE30 vector and pQEMgtR plasmid were used as control.
Role of MgtR in intramacrophage survival
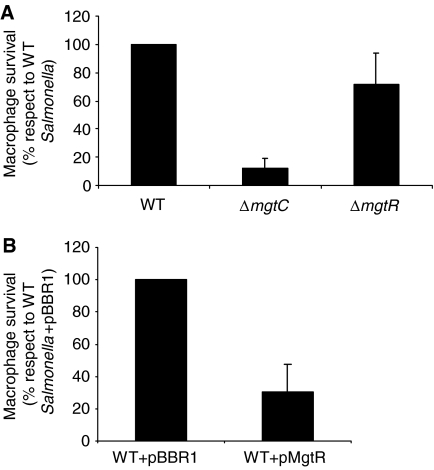
MgtC plays a role for Salmonella intramacrophage growth (Blanc-Potard and Groisman, 1997). The mutations E84A, G85A and N92T in MgtC produce proteins that can not complement the macrophage growth defect of a S. typhimurium ΔmgtC mutant (Rang et al, 2007; data not shown). We have compared the intramacrophage survival of a S. typhimurium wild-type strain and a ΔmgtR-mutant strain. The absence of MgtR had only a slight effect on intracellular growth (Figure 10A), indicating that the stabilization of MgtC has no major effect in this virulence assay. In addition, the ΔmgtR-mutant strain grew similarly as the wild-type strain in low-Mg2+ medium (data not shown). On the other hand, we have shown that the overexpression of MgtR in an S. typhimurium wild-type strain significantly reduced the intramacrophage growth (Figure 10B). This intracellular growth defect is not linked to a slow down of bacterial growth rate, since strains exhibited similar growth curves in Luria Broth (LB) medium or low-Mg2+ medium (data not shown).
Figure 10.
Role of MgtR in S. typhimurium intramacrophage growth. The replication of Salmonella strains in J774 macrophages was evaluated 18 h after infection. Data represent the mean values plus standard errors from at least three independent experiments. (A) Analysis of a ΔmgtR-mutant strain in comparison with a wild-type strain and a ΔmgtC mutant. Values presented are the percentage relative to that of the wild-type S. typhimurium 14028s. (B) Analysis of a wild-type Salmonella strain that carries the pMgtR plasmid. Values presented are the percentage relative to that of the wild-type S. typhimurium 14028s with the pBBR1MCS vector.
Discussion
Multiple regulatory pathways control the MgtC protein levels
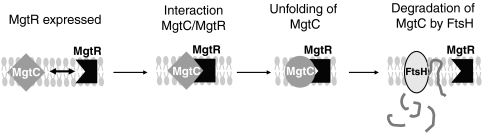
The mgtCB operon is known to be regulated at the transcriptional level by the PhoPQ two-component system in response to external Mg2+ concentration (García-Véscovi et al, 1996). The PhoP regulatory protein interacts directly with the mgtCB promoter (Zwir et al, 2005). In addition, the operon is regulated at the transcriptional level by intracellular Mg2+ concentration through a PhoPQ-independent mechanism that depends on the secondary structure of its 5′UTR (Groisman et al, 2006). In the present study, we describe an additional level of regulation at the post-translational level for the MgtC protein. Expression of the MgtC protein is prevented by the MgtR peptide, which is encoded downstream of the mgtB gene. The MgtR peptide directly interacts with MgtC and probably unfolds MgtC to promote its degradation by the membrane-bound AAA+ protease FtsH. This novel regulatory mechanism, which identifies for the first time a virulence factor as FtsH target, is schematized in Figure 11.
Figure 11.
Model for the role of MgtR in MgtC expression: the MgtR peptide binds directly to MgtC at the cytoplasmic membrane. The interaction between MgtC and MgtR would unfold MgtC and make it a target for degradation by the FtsH protease.
MgtR, a regulatory peptide that interacts with MgtC and promotes its degradation by the FtsH protease
MgtC is an inner-membrane protein that harbours five transmembrane segments in the first half of the protein (Rang et al, 2007). MgtR is an inner-membrane peptide with a single highly hydrophobic transmembrane domain. Using the bacterial two-hybrid BACHT system, which appears as the technique of choice to study interaction between bacterial proteins within biological membranes (Karimova et al, 2005; Schneider et al, 2007), we could demonstrate interaction between MgtC and MgtR. The strength of this interaction correlates with the ability of MgtR to mediate MgtC degradation. The hydrophobicity and the α-helical structure of MgtR are important for its function. The interaction between membrane-imbedded helices remains hard to predict solely on the basis of amino-acid sequences, but some motifs can drive helix–helix interactions (Schneider et al, 2007). MgtR harbours an Ala-coil motif that has been implicated in the association of membrane-imbedded helices (Walters and DeGrado, 2006) This Ala-coil motif appears essential for MgtC–MgtR interaction and MgtR-mediated MgtC degradation. We identified mutations in MgtR, as S17I or A24L, that do not prevent the expression or membrane location of the peptide, and completely abolish its regulatory function and interaction with MgtC. We have also identified mutations in MgtC that prevented its downregulation by MgtR and lowered the interaction of MgtC with MgtR in the BACHT system. These mutations are located in a cytoplasmic loop between the third and fourth transmembrane domains (TM3 and TM4) of MgtC. The context of a transmembrane helix is essential for its correct topology and loops can contribute to the stability of helix interactions (Schneider et al, 2007). Hence, loop mutations could disrupt the helical packing in TM3 and/or TM4 and thereby prevent interaction with MgtR. Interestingly, the fourth helical segment of MgtC harbours an Ala-coil motif. However, mutagenesis of single residues did not indicate a contribution of this motif in the interaction with MgtR. The interacting domain of MgtC might be complex and involve more than a single TM domain.
A kinetic experiment demonstrated that MgtR promotes MgtC degradation, although through a relatively slow process. MgtR acts specifically on MgtC since no effect was detected on MgtB. The fact that both MgtC and MgtR are encoded by the same operon is rather surprising and raises the question of timing for MgtC expression and the regulatory peptide expression. Previous studies have indicated that MgtC can be detected after limited time of Mg2+ starvation (2–4 h) (Moncrief and Maguire, 1998; Adkins et al, 2006; data not shown), but MgtC is not expressed anymore after 16 h Mg2+ starvation (Moncrief and Maguire, 1998; Figure 3A). Our results suggest that upon induction of the mgtCBR operon, MgtR does not affect MgtC production, MgtC being similarly expressed in the presence or absence of MgtR after short time induction (data not shown), but that the production of MgtR promotes MgtC degradation after few hours.
We have shown that the degradation of MgtC is dependent on the presence of a functional FtsH protease. FtsH is the only membrane-bound AAA+ protease in E. coli and this protease plays an important role in the quality control of membrane proteins (Ito and Akiyama, 2005). We propose that the interaction between MgtC and MgtR might unfold MgtC and make it a substrate for FtsH-mediated degradation. FtsH recognizes unstructured peptide sequences, which are typically located at the N- or C-terminal end of a target protein, and degrades target protein processively. Internal loops also contribute to the degradation of substrates by FtsH and other AAA+ proteases (Datta et al, 2005; Hoskins and Wickner, 2006; Okuno et al, 2006). To date, the precise mechanism of degradation of MgtC by FtsH remains unknown.
To our knowledge, MgtR is the first example of an α-helical hydrophobic peptide that modulates degradation of a bacterial membrane protein by an AAA+ protease. It is possible that the participation of peptides in the regulation of protein stability might be a more common process that has not been characterized yet due to the lack of appropriate screening/detection methods. To date, the only other reported case of a peptide that modulates AAA+ protease activity is the SpoVM peptide from Bacillus subtilis, which, in contrast to MgtR, is an amphipathic helix (Cutting et al, 1997; Prajapati et al, 2000). SpoVM has been involved in sporulation and has an inhibitory action on the FtsH protease. In addition, several adaptor proteins modulate proteolysis by enhancing or inhibiting interaction between specific substrates and AAA+ proteases (Dougan et al, 2002; Baker and Sauer, 2006). Most adaptor proteins interact directly with the substrate in complex with the AAA+ partner protein and they usually modulate recognition of the substrate. Adaptors are generally small proteins encoded by genes that are rarely co-transcribed with the genes encoding their target(s). MgtR differs from previously described adaptors in its size and coexpression with its target.
Physiological role of MgtR during infection
MgtR is conserved in other Salmonella enterica serovars (S. typhi and S. choleraesuis), but is not found in other bacterial species that encode MgtC. Hence, it seems that Salmonella has developed a specific regulatory system that acts as a negative feedback to limit the amount of MgtC protein. We propose that this feedback regulation is linked to the high transcriptional induction of the Salmonella mgtC gene under certain conditions. In other bacteria, the mgtC gene might be expressed at lower levels, preventing the need for a feedback regulatory mechanism. MgtC has likely been acquired by horizontal gene transfer and is known to be differentially regulated in various species (Alix and Blanc-Potard, 2007). MgtC is required for intramacrophage growth (Alix and Blanc-Potard, 2007) and the Salmonella mgtCB operon is highly induced in macrophages (Smith et al, 1998; Eriksson et al, 2003). However, a proteomic analysis of S. typhimurium isolated from macrophages identified MgtB but not MgtC (Shi et al, 2006). This suggests that the intracellular level of MgtC protein is low and/or that there is a detectable expression at infection times that were not experimentally tested. To better understand the physiological role of MgtR during Salmonella infection, we have investigated the growth phenotype of a ΔmgtR mutant in macrophages. The ΔmgtR mutant is only slightly attenuated for growth within macrophages, indicating that MgtR does not play a major role in this virulence assay. On the other hand, overexpression of MgtR in a wild-type strain of S. typhimurium reduces significantly the ability of the strain to grow within macrophages. This might be linked to a high degradation level of the MgtC virulence factor. Hence, this result suggests that MgtR could act as an antagonist of the MgtC virulence factor under certain conditions.
Conclusion
The completion of bacterial genomes has significantly improved the description of regulatory networks. However, small ORFs are not annotated on genome maps. In addition, functional small ORFs are rarely detected by genetic screening due to the low frequency of mutational or insertion events in these regions. To date, studies on small ORFs have mainly focused on the identification of small regulatory RNAs. In the present study, we have identified a regulatory peptide involved in the degradation of a virulence factor. In Salmonella, a recent peptidomics analysis, which did not include peptides encoded by non-annotated ORFs, has suggested a targeted protein degradation under phagosome-mimicking culture conditions (Manes et al, 2007). Further peptidomics research is required to characterize additional functional bacterial peptides, which can be encoded by small ORFs or can be generated by the proteolytic cleavage of proteins.
Materials and methods
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in Table I. All S. typhimurium strains are derived from wild-type 14028s with the exception of TH6767 and NM643 that are derived from LT2. E. coli thermosensitive ftsH1 mutant (Begg et al, 1992) is derived from W3110 and has been provided by T Ogura. E. coli DH5α (Hanahan, 1983) was used as host in cloning experiments. Bacteria were grown in LB supplemented with suitable antibiotics to maintain plasmid DNA (ampicillin (Amp) at 100 μg ml−1, kanamycin (Kan) at 25 μg ml−1, chloramphenicol (Cm) at 10 μg ml−1). Growth in low-Mg2+ liquid medium was carried out in NCE-minimal medium (Maloy, 1990) supplemented with 0.1% casamino acids, 38 mM glycerol and 10 μM MgCl2. Growth in high-Mg2+ liquid medium was carried out in the same medium supplemented with 10 mM MgCl2.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description or phenotype | Reference or source |
|---|---|---|
| S. typhimurium | ||
| 14028s | Wild type | EA Groisman |
| MA6987 | ilvI3305∷Tn10dTac-cat/pKD46 | Uzzau et al (2001) |
| TH6767 | clpX1∷Tn10dcam | Aldridge et al (2003) |
| NM14 | ΔmgtC | Rang et al (2007) |
| NM506 | ΔmgtR∷kan | This study |
| NM516 | ΔmgtR | This study |
| NM643 | clpX1∷Tn10dcam ΔmgtR∷kan | This study |
| E. coli | ||
| DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rk−, mk+) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| AR3307 | W3110 zad220∷Tn10 | Ogura et al (1999) |
| AR3317 | AR3307 zgj3198∷Tn10 (kanR) ftsH1(Ts) | Ogura et al (1999) |
| BTH101 | F− cya-99 araD139 galE15 galK16 rpsL1 (Strr) hsdR2 mcrA1 mcrB1 | D Ladant |
| Cloning plasmids | ||
| pBR322 | AmpR TcR reppMB1 | Bolivar et al (1977) |
| pBBR1MCS | CmR | Kovach et al (1994) |
| pQE30 | AmpR repColE1 | Qiagen |
| pUT18 | AmpR repColE1 (pBluescript II KS derivative) | Karimova et al (1998) |
| pKT25ΔXbaI | KanR repp15A (pACYC184 derivative) | G Patey |
| pREP4 | KanR repp15A lacIq | Qiagen |
| Plasmids | ||
| pEG9094 | pBR322 mgtC+ | Blanc-Potard and Groisman (1997) |
| pEG9091 | pBR322 mgtC+ mgtB+ mgtR+ | Blanc-Potard and Groisman (1997) |
| pEG9091 del1 | pEG9091 mgtC+ mgtBΔ439–904 mgtR+ | This study |
| pEG9091 del2 | pEG9091 mgtC+ mgtBΔ235–904 mgtR+ | This study |
| pEG9091 del3 | pEG9091 mgtC+ mgtBΔ60–904 mgtR+ | This study |
| pEG9091 del4 | pEG9091 mgtC+ mgtB+ | This study |
| pEG9091 del5 | pEG9091 mgtC+ mgtBΔ60–910 | This study |
| pNM12 | pBR322 mgtC+ | Rang et al (2007) |
| pMgtR | pBBR1MCS mgtR+ | This study |
| pQEMgtR | His6-MgtR | This study |
| pUT18-MgtC | MgtC-T18 | This study |
| pKT25ΔXbaI-MgtR | T25-MgtR | This study |
Plasmids construction
Plasmids purified with Qiaprep spin kit (Qiagen) were introduced into S. typhimurium strains by electroporation using a Bio-Rad apparatus, and into E. coli strains by chemical transformation using standard procedures. Plasmids used in this study are listed in Table I, and primers are listed in Supplementary Table S1.
Deletions of sequences downstream of mgtC were carried out on the pEG9091 plasmid that harbours the mgtCB operon (Figure 1A). PCR amplification was performed with primers flanking the region to delete. Amplified fragments were phosphorylated and self-ligated.
To clone mgtR, a 1200-bp HindIII–PstI restriction fragment from pEG9091del3, which carries a 270-bp region downstream of mgtB, was first subcloned into pBBR1MCS, which is a CmR vector compatible with pBR322 derivatives. Deletions were subsequently made at the 3′ (with primers sup3′-F and sup3′-R) and 5′ (with primers sup5′-F and MgtB-del-F) extremities of the insert to keep a 150-bp region downstream of mgtB that includes the last six codons of mgtB. The resulting plasmid was called pMgtR.
To produce a His-tagged MgtR peptide, we cloned the mgtR gene between the BamHI and HindIII sites of the pQE30 vector (Qiagen). The cloned fragment resulted from the annealing of long oligonucleotides (MgtR-BamHind-F and MgtR-HindBam-R) corresponding to the entire mgtR sequence with cloning sites at the ends. The resulting plasmid, pQEMgtR, encodes an N-terminal His-tagged MgtR. Overexpression of MgtR was not well tolerated by bacteria and pQEMgtR was maintained in strains that harbour the lacIq gene on pREP4 plasmid. Expression of His6-MgtR was induced by adding 0.1 mM IPTG.
Site-directed mutagenesis was conducted on plasmids using the Quickchange® II kit (Stratagene) according to the manufacturer's instructions. Sets of primers were designed for each engineered mutation including sufficiently long flanking regions. Sequencing was conducted to confirm the presence of the desired mutation and the absence of additional mutations.
Western blot analysis
Bacteria were grown overnight in low-Mg2+ medium (to induce mgtC transcription) supplemented with suitable antibiotics to maintain plasmid DNA. To prepare whole-cell extracts, cultures were normalized for the number of cells, centrifuged, resuspended in 100 μl Laemmli buffer and lysed by 10 min boiling. Samples were run on 12 % SDS–PAGE gel and transferred to PVDF membrane (Millipore) for immunoblotting. Anti-MgtC antibodies were raised in rabbits against the His-tagged C-terminal domain of the MgtC protein (Rang et al, 2007). Hence, these antibodies react both with MgtC protein and His-tagged proteins. Mouse anti-DnaK antibodies (Tebubio) used at 1:5000 dilution were used as control. Rabbit anti-MgtB antibodies, kindly provided by M Maguire, were used at 1:1000 dilution. Secondary horseradish peroxidase-conjugated anti-rabbit or anti-mouse antiserum (Sigma) were used at 1:3000 dilution. The blots were developed with the Immobilon™ Western kit (Millipore).
Construction of a ΔmgtR Salmonella strain
A chromosomal deletion in the mgtR gene was constructed using λRed-mediated site-specific recombination (Datsenko and Wanner, 2000). The kanamycin-resistance gene of plasmid pKD13 (Datsenko and Wanner, 2000) was amplified by PCR using primers Δ-MgtR-F and Δ-MgtR-R. The resulting PCR product was used to replace 76 bp of mgtR in strain MA6897 (Uzzau et al, 2001) by homologous recombination catalysed by the λRed recombinase encoded by plasmid pKD46. The kanamycin-resistance cassette inserted at the chromosomal mgtR locus was transferred by P22 transduction into a wild-type 14028s strain. The temperature-sensitive plasmid pCP20 encoding FLP recombinase (Datsenko and Wanner, 2000) was transformed in the resulting strain to allow loss of the kanamycin resistance cassette, leaving behind a single FRT site. The chromosomal deletion was verified by colony PCR using primers MgtB-del-F and MgtRseqR followed by sequencing.
RNA extraction and RT–PCR
Overnight culture of wild-type 14028s strain was diluted 1:50 into 3 ml of NCE medium containing 10 mM Mg2+ and grown at 37°C until OD600 reached a value of 0.7. Bacteria were harvested by centrifugation, washed three times with NCE medium containing 10 μM Mg2+ and cultivated 30 min at 37°C in 3 ml of the same medium before RNA extraction. Total RNA was isolated using the SV Total RNA Isolation System (Promega) and treated with RNase-free DNase I following the protocol provided by the manufacturer. RT–PCRs were performed with the AccessQuick™ RT–PCR System (Promega) following the manufacturer's instructions. Primers used are RT-GapA-F and RT-GapA-R for gapA, MgtC-RT-F2 and MgtC-RT-R2 for amplification 1, MgtR-RT-F and MgtR-RT-R for amplification 2, MgtB-del-F and MgtR-RT-R for amplification 3, MgtBR-RT-F and MgtR-RT-R for amplification 4. Reverse transcription was performed by 45-min incubation at 45°C, and PCR was performed as follows: 2-min initial denaturation at 95°C, 28 repetitions of 30 s at 95°C, 30 s at 50°C and 30 s at 72°C, and a final amplification of 5 min at 72°C. To exclude possible DNA contamination, the same reactions were performed without reverse transcriptase.
Bacterial two-hybrid analysis
We used the BACTH system (Karimova et al, 1998). The mgtC gene was PCR amplified using pEG9091 as template and primers MgtC-pUT18-F and MgtC-pUT18-R. The PCR fragment was cloned at the SmaI site of the pUT18 vector, to produce a fusion protein MgtC-T18. Expression of MgtC-T18 hybrid protein and mutant derivatives was checked by Western analysis. The pKT25 vector (Karimova et al, 1998) was modified by deleting the XbaI site to produce pKT25ΔXbaI that allows in frame cloning at the BamHI site. The mgtR gene was amplified by colony PCR from 14028s with Pfu polymerase (Promega) using primers MgtR-pKT25-F and MgtR-pKT25-R. The blunt-ended PCR fragment was digested by BamHI and cloned into the pKT25ΔXbaI vector, previously digested by EcoRI, treated with Klenow and digested by BamHI, to produce a fusion protein T25-MgtR.
Recombinant plasmids carrying mgtC and mgtR genes were co-transformed into BTH101 bacteria. Transformants were plated on LB Amp Kan X-gal medium at 30°C for 30 h. To quantify the interaction between hybrid proteins, bacteria were grown overnight either at 30°C (as recommended in the BACHT protocol) or at 37°C (where interaction is usually weaker comparatively to 30°C) in LB Amp Kan liquid medium supplemented with 0.5 mM IPTG. Before the β-galactosidase assay, cultures were diluted 1:3 in LB medium. β-Galactosidase assays were carried out as described previously (Miller, 1972), and activities are expressed in arbitrary Miller units. Values are average from at least six independent cultures. A level of β-galactosidase activity at least fivefold higher than that measured for vectors indicates an interaction.
Macrophage infection experiments
The rate of intramacrophage replication after 18 h infection was performed in J774 mouse macrophages as described previously (Rang et al, 2007).
Accession number
The mgtR sequence has been submitted to GenBank database under accession number EU154350.
Supplementary Material
Supplementary Information
Supplementary Table S1
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Acknowledgments
We thank Lionello Bossi (Gif-sur Yvette, France), Eduardo A Groisman (St Louis, MO), Kelly T Hughes (Salt Lake City, UT), Daniel Ladant (Paris, France), Gilles Patey and Teru Ogura (Kumamoto, Japan) for providing bacterial strains and plasmids; Michael E Maguire for providing antibodies; Gregory Baronian for help in the mutagenesis of MgtR and Cécile Rang for RNA extraction; L Bossi and EA Groisman for critical reading of the paper and Gilles Labesse (Montpellier, France) and David O'Callaghan for helpful discussions. This work was supported by Inserm (Avenir program), Université Montpellier 1, la région Languedoc-Roussillon and la ville de Nîmes. EA is supported by Inserm and Région Languedoc-Roussillon.
References
- Adkins JN, Mottaz HM, Norbeck AD, Gustin JK, Rue J, Clauss TR, Purvine SO, Rodland KD, Heffron F, Smith RD (2006) Analysis of the Salmonella typhimurium proteome through environmental response toward infectious conditions. Mol Cell Proteomic 5: 1450–1461 [DOI] [PubMed] [Google Scholar]
- Aldridge P, Karlinsey J, Hughes KT (2003) The type III secretion chaperone FlgN regulates flagellar assembly via a negative feedback loop containing its chaperone substrates FlgK and FlgL. Mol Microbiol 49: 1333–1345 [DOI] [PubMed] [Google Scholar]
- Alix E, Blanc-Potard AB (2007) MgtC: a key player in intramacrophage survival. Trends Microbiol 15: 252–256 [DOI] [PubMed] [Google Scholar]
- Baker TA, Sauer RT (2006) ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem Sci 31: 647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg KJ, Tomoyasu T, Donachie WD, Khattar M, Niki H, Yamanaka K, Hiraga S, Ogura T (1992) Escherichia coli mutant Y16 is a double mutant carrying thermosensitive ftsH and ftsI mutations. J Bacteriol 174: 2416–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc-Potard AB, Groisman EA (1997) The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J 16: 5376–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc-Potard AB, Lafay B (2003) MgtC as a horizontally acquired virulence factor of intracellular bacterial pathogens: evidence from molecular phylogeny and comparative genomics. J Mol Evol 57: 479–486 [DOI] [PubMed] [Google Scholar]
- Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW (1977) Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2: 95–113 [PubMed] [Google Scholar]
- Buchmeier N, Blanc-Potard A, Ehrt S, Piddington D, Riley L, Groisman EA (2000) A parallel intraphagosomal survival strategy shared by Mycobacterium tuberculosis and Salmonella enterica. Mol Microbiol 35: 1375–1382 [DOI] [PubMed] [Google Scholar]
- Cutting S, Anderson M, Lysenko E, Page A, Tomoyasu T, Tatematsu K, Tatsuta T, Kroos L, Ogura T (1997) SpoVM, a small protein essential to development in Bacillus subtilis, interacts with the ATP-dependent protease FtsH. J Bacteriol 179: 5534–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta AB, Roy S, Parrack P (2005) Role of C-terminal residues in oligomerization and stability of lambda CII: implications for lysis–lysogeny decision of the phage. J Mol Biol 345: 315–324 [DOI] [PubMed] [Google Scholar]
- Dougan DA, Mogk A, Zeth K, Turgay K, Bukau B (2002) AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett 529: 6–10 [DOI] [PubMed] [Google Scholar]
- Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC (2003) Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47: 103–118 [DOI] [PubMed] [Google Scholar]
- Frees D, Qazi SN, Hill PJ, Ingmer H (2003) Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol Microbiol 48: 1565–1578 [DOI] [PubMed] [Google Scholar]
- García-Véscovi E, Soncini FC, Groisman EA (1996) Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84: 165–174 [DOI] [PubMed] [Google Scholar]
- Gottesman S (2003) Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol 19: 565–587 [DOI] [PubMed] [Google Scholar]
- Grabenstein JP, Fukuto HS, Palmer LE, Bliska JB (2006) Characterization of phagosome trafficking and identification of PhoP-regulated genes important for survival of Yersinia pestis in macrophages. Infect Immun 74: 3727–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA, Cromie MJ, Shi Y, Latifi T (2006) A Mg2+-responding RNA that controls the expression of a Mg2+ transporter. Cold Spring Harb Symp Quant Biol 71: 251–258 [DOI] [PubMed] [Google Scholar]
- Günzel D, Kucharski LM, Kehres DG, Romero MF, Maguire ME (2006) The MgtC virulence factor of Salmonella enterica serovar Typhimurium activates Na+,K+-ATPase. J Bacteriol 188: 5586–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166: 557–580 [DOI] [PubMed] [Google Scholar]
- Hoskins JR, Wickner S (2006) Two peptide sequences can function cooperatively to facilitate binding and unfolding by ClpA and degradation by ClpAP. Proc Natl Acad Sci USA 103: 909–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Akiyama Y (2005) Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol 59: 211–231 [DOI] [PubMed] [Google Scholar]
- Karimova G, Dautin N, Ladant D (2005) Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 187: 2233–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA 95: 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Phillips RW, Elzer PH, Roop RM, Peterson KM (1994) pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16: 800–802 [PubMed] [Google Scholar]
- Kwon HY, Ogunniyi AD, Choi MH, Pyo SN, Rhee DK, Paton JC (2004) The ClpP protease of Streptococcus pneumoniae modulates virulence gene expression and protects against fatal pneumococcal challenge. Infect Immun 72: 5646–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigne JP, O'callaghan D, Blanc-Potard AB (2005) Requirement of MgtC for Brucella suis intramacrophage growth: a potential mechanism shared by Salmonella enterica and Mycobacterium tuberculosis for adaptation to a low-Mg2+ environment. Infect Immun 73: 3160–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM (2006) Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog 2: e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney KE, Valvano MA (2006) The mgtC gene of Burkholderia cenocepacia is required for growth under magnesium limitation conditions and intracellular survival in macrophages. Infect Immun 74: 5477–5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy SR (1990) Experimental Techniques in Bacterial Genetics. Boston, MA: Jones and Bartlett Publishers [Google Scholar]
- Manes NP, Gustin JK, Rue J, Mottaz HM, Purvine SO, Norbeck AD, Monroe ME, Zimmer JS, Metz TO, Adkins JN, Smith RD, Heffron F (2007) Targeted protein degradation by Salmonella under phagosome-mimicking culture conditions investigated using comparative peptidomics. Mol Cell Proteomics 6: 717–727 [DOI] [PubMed] [Google Scholar]
- Miller JH (1972) Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- Moncrief MB, Maguire ME (1998) Magnesium and the role of MgtC in growth of Salmonella typhimurium. Infect Immun 66: 3802–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Inoue K, Tatsuta T, Suzaki T, Karata K, Young K, Su LH, Fierke CA, Jackman JE, Raetz CR, Coleman J, Tomoyasu T, Matsuzawa H (1999) Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol Microbiol 31: 833–844 [DOI] [PubMed] [Google Scholar]
- Okuno T, Yamanaka K, Ogura T (2006) An AAA protease FtsH can initiate proteolysis from internal sites of a model substrate, apo-flavodoxin. Genes Cells 11: 261–268 [DOI] [PubMed] [Google Scholar]
- Prajapati RS, Ogura T, Cutting SM (2000) Structural and functional studies on an FtsH inhibitor from Bacillus subtilis. Biochim Biophys Acta 1475: 353–359 [DOI] [PubMed] [Google Scholar]
- Rang C, Alix E, Felix C, Heitz A, Tasse L, Blanc-Potard AB (2007) Dual role of the MgtC virulence factor in host and non-host environments. Mol Microbiol 63: 605–622 [DOI] [PubMed] [Google Scholar]
- Schneider D, Finger C, Prodohl A, Volkmer T (2007) From interactions of single transmembrane helices to folding of alpha-helical membrane proteins: analyzing transmembrane helix–helix interactions in bacteria. Curr Protein Pept Sci 8: 45–61 [DOI] [PubMed] [Google Scholar]
- Shi L, Adkins JN, Coleman JR, Schepmoes AA, Dohnkova A, Mottaz HM, Norbeck AD, Purvine SO, Manes NP, Smallwood HS, Wang H, Forbes J, Gros P, Uzzau S, Rodland KD, Heffron F, Smith RD, Squier TC (2006) Proteomic analysis of Salmonella enterica serovar Typhimurium isolated from RAW 264.7 macrophages: identification of a novel protein that contributes to the replication of serovar Typhimurium inside macrophages. J Biol Chem 281: 29131–29140 [DOI] [PubMed] [Google Scholar]
- Smith RL, Kaczmarek MT, Kucharski LM, Maguire ME (1998) Magnesium transport in Salmonella typhimurium: regulation of mgtA and mgtCB during invasion of epithelial and macrophage cells. Microbiology 144: 1835–1843 [DOI] [PubMed] [Google Scholar]
- Snavely MD, Miller CG, Maguire ME (1991) The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J Biol Chem 266: 815–823 [PubMed] [Google Scholar]
- Takaya A, Kubota Y, Isogai E, Yamamoto T (2005) Degradation of the HilC and HilD regulator proteins by ATP-dependent Lon protease leads to downregulation of Salmonella pathogenicity island 1 gene expression. Mol Microbiol 55: 839–852 [DOI] [PubMed] [Google Scholar]
- Takaya A, Suzuki M, Matsui H, Tomoyasu T, Sashinami H, Nakane A, Yamamoto T (2003) Lon, a stress-induced ATP-dependent protease, is critically important for systemic Salmonella enterica serovar typhimurium infection of mice. Infect Immun 71: 690–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L (2001) Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci USA 98: 15264–15269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RF, DeGrado WF (2006) Helix-packing motifs in membrane proteins. Proc Natl Acad Sci USA 103: 13658–13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Sashinami H, Takaya A, Tomoyasu T, Matsui H, Kikuchi Y, Hanawa T, Kamiya S, Nakane A (2001) Disruption of the genes for ClpXP protease in Salmonella enterica serovar Typhimurium results in persistent infection in mice, and development of persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect Immun 69: 3164–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwir I, Shin D, Kato A, Nishino K, Latifi T, Solomon F, Hare JM, Huang H, Groisman EA (2005) Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci USA 102: 2862–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Table S1
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4