SUMMARY
Eukaryotic mRNAs harboring premature translation termination codons are recognized and rapidly degraded by the nonsense-mediated mRNA decay (NMD) pathway. The mechanism for discriminating between mRNAs that terminate translation prematurely and those subject to termination at natural stop codons remains unclear. Studies in multiple organisms indicate that proximity of the termination codon to the 3′ poly(A) tail and the poly(A) RNA binding protein, PAB1, constitute the critical determinant in NMD substrate recognition. We demonstrate that mRNA in yeast lacking a poly(A) tail can be destabilized by introduction of a premature termination codon and importantly, this mRNA is a substrate of the NMD machinery. We further show that in cells lacking Pab1p, mRNA substrate recognition and destabilization by NMD is intact. These results establish that neither the poly(A) tail, nor PAB1, is required in yeast for discrimination of nonsense codon-containing mRNA from normal by NMD.
INTRODUCTION
Nonsense-mediated mRNA decay (NMD) is a conserved quality control mechanism in eukaryotic cells that recognizes and eliminates mRNA encoding premature translational termination signals in their protein coding region (Baker and Parker, 2004; Maquat, 2004). The rapid degradation of mRNA lacking a complete open reading frame (ORF) protects cells from accumulation of truncated polypeptides that can confer deleterious dominant-negative or gain-of-function phenotypes (Pulak and Anderson, 1993). Mutations in components of the NMD machinery lead to mental retardation in humans or embryonic lethality in mice, underscoring the functional importance of NMD (Medghalchi et al, 2001; Tarpey et al, 2007). Furthermore, NMD is a potent modulator of phenotypic outcome for many human genetic diseases (Frischmeyer and Dietz, 1999; Kuzmiak and Maquat, 2006).
NMD substrates encompass not only mRNAs harboring premature termination codons (PTCs) introduced by mutation, but also a significant population of the yeast, Drosophila, and human transcriptomes (He et al, 2003; Lelivelt and Culbertson, 1999; Mendell et al, 2004; Rehwinkel et al, 2005). Many endogenous mRNA targets that undergo NMD contain PTCs as a consequence of inefficient pre-mRNA splicing (He et al, 1993), upstream ORFs (Oliveira and McCarthy, 1995), or use of out-of-frame AUG start codons (Welch and Jacobson, 1999). Interestingly, mRNAs terminating translation at a natural stop codon but harboring an extension of their 3′ UTR are also substrates for NMD (Muhlrad and Parker, 1999; Pulak and Anderson, 1993). For all NMD substrates, therefore, translation termination occurs in an inappropriate context and NMD appears to result from an alteration in the spatial relationship between the termination codon and other features of the mRNA (Baker and Parker, 2004).
Translation termination is considered aberrant as a consequence of its occurrence upstream of a second signal consisting of mRNA sequence elements and associated protein markers. In one model, prematurely terminating ribosomes fail to displace a downstream mRNA-protein complex (mRNP) able to trigger decay of the mRNA by recruiting proteins required for NMD (Maquat, 2004). Ribosomes that elongate through the entire ORF and terminate at a natural stop codon, in contrast, displace the downstream mRNP to prevent NMD. Proteins deposited at exon-exon junctions (exon junction complexes, EJCs) during pre-mRNA splicing serve to mark upstream stop codons as premature in mammalian cells (Zhang et al, 1998a; 1998b). Although recognition of mRNAs harboring PTCs is highly dependent upon a downstream exon-exon junction in vertebrates, exceptions have been described, demonstrating that a downstream EJC is not universally required for mammalian NMD (Buhler et al, 2006; Wang et al, 2002; Weil and Beemon, 2006).
In yeast, flies, worms, and plants, PTC definition can occur independently of a downstream exon boundary, and EJC protein components are either predominantly absent or not required for NMD (Culbertson and Leeds, 2003; Gatfield et al, 2003; Longman et al, 2007; Muhlrad and Parker, 1994; van Hoof and Green, 1996). Therefore, other determinants must present the downstream signal required for NMD substrate recognition. Yeast mRNAs are proposed to harbor downstream sequence elements (DSEs) within the coding region which bind protein factors (e.g. Hrp1) that trigger NMD if not removed during translation elongation (Gonzalez et al, 2000; Peltz et al, 1993). The few characterized DSEs, however, share loose sequence consensus making it difficult to estimate their ubiquity within ORFs and their importance in NMD. Moreover, analogous elements have not been identified in other organisms (Behm-Ansmant et al, 2007; Maquat, 2004).
An alternative paradigm for recognition of nonsense-containing mRNAs is the ‘faux UTR’ model which posits that translation termination at a PTC is intrinsically aberrant because it occurs upstream of elements unable to function as an authentic 3′ UTR (Amrani et al, 2004, 2006). Ribosomes at a normal stop codon, in contrast, terminate proximal to the 3′ UTR and engage in interactions necessary for efficient termination and stabilization of the mRNA. According to this model, an appropriate mRNP context includes the mRNA poly(A) tail and poly(A) binding protein, PAB1. Importantly, tethering of PAB1 downstream and proximal to a PTC stabilizes NMD substrates in both yeast and Drosophila (Amrani et al, 2004; Behm-Ansmant et al, 2007). In addition, targeting of mRNAs with extended 3′ UTRs by NMD is consistent with PAB1 and the poly(A) tail providing important positional information for substrate recognition by NMD.
We evaluated the requirement for an mRNA poly(A) tail and PAB1 in recognition of NMD substrates. We show that introduction of a PTC into a yeast mRNA lacking a poly(A) tail can result in destabilization of the mRNA, and that rapid decay of the mRNA requires the NMD factor Upf1p, confirming it as a bona fide NMD substrate. Further, we reveal that in the absence of Pab1p, mRNAs harboring a PTC are recognized and destabilized in an Upf1p-dependent manner. Our findings clearly demonstrate that in yeast, neither an mRNA poly(A) tail nor Pab1p is required for NMD. These findings illustrate that the proximity of the terminating ribosome to PAB1 either plays no role in substrate discrimination by NMD, or is just one of several redundant features that distinguish a termination event as premature.
RESULTS
Unadenylated GFP-RZ mRNA is Targeted for Degradation by the Cytoplasmic Exosome
To evaluate the relevance of the poly(A) tail in NMD in yeast, we utilized a reporter gene containing the GFP ORF followed immediately by a hammerhead ribozyme (GAL1-GFP-RZ; Figure 1A; Dower et al, 2004). Inclusion of the ribozyme sequence results in RNA cleavage that generates a 6 nucleotide 3′ UTR that fails to undergo polyadenylation (see Supplemental Material).
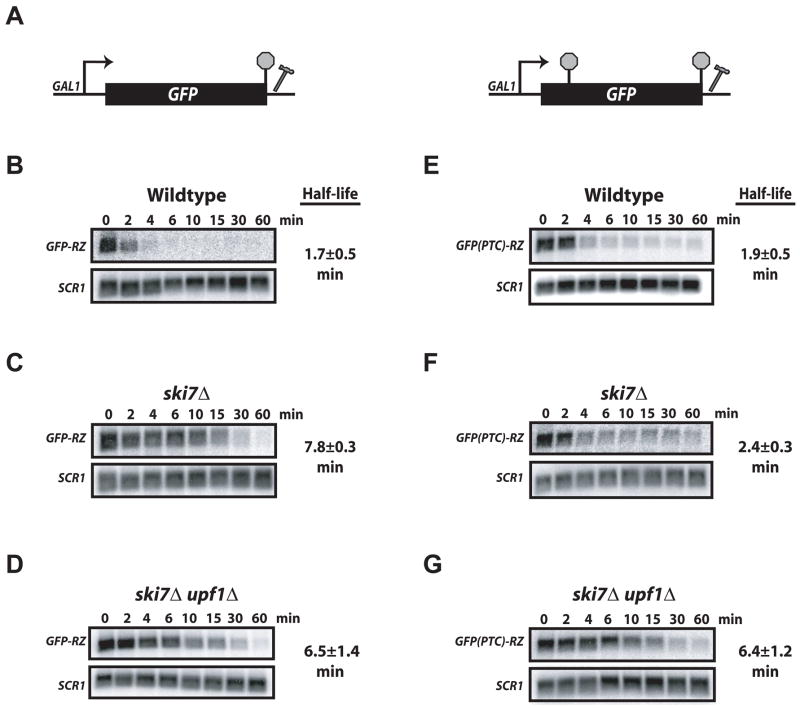
Figure 1. Unadenylated GFP(PTC)-RZ mRNA is Targeted for NMD.
A. Schematic of GFP-RZ and GFP(PTC)-RZ reporter genes. Natural and premature stop codons (stop signs) and location of the hammerhead ribozyme directly 3′ of the GFP ORF are depicted. GFP-RZ and GFP(PTC)-RZ mRNA stability was determined by transcriptional shut-off analysis in wild-type cells (B&E), a mutant lacking cytoplasmic exosome activity, ski7Δ (C&F), and cells lacking both Ski7p and Upf1p (D&G). RNA isolated at various times after inhibition of reporter gene transcription (min) was analyzed by Northern blot and probed for GFP mRNA and SCR1 RNA (loading control). mRNA half-lives are the average ± standard deviation of four independent experiments.
Meaux and van Hoof (2006) reported that unadenylated mRNAs are rapidly degraded by the cytoplasmic exosome. To evaluate if GFP-RZ mRNA is also a substrate for the exosome, transcript stability was determined in wild-type cells and a strain lacking cytoplasmic exosome activity (ski7Δ). This analysis revealed that GFP-RZ mRNA was unstable in wild-type cells decaying with a half-life of approx. 2 min (Figure 1B; Supplemental Figure 1). In contrast, GFP-RZ mRNA was stable in ski7Δ cells, with a half-life of 8 min (Figure 1C). Unadenylated GFP-RZ mRNA is therefore present in the cytoplasm where it is degraded by the cytoplasmic exosome.
Nonsense Codon-containing Unadenylated mRNA is Targeted for Rapid Degradation
It has been shown that GFP-RZ mRNA levels are low, and not further decreased by the introduction of a PTC (Baker and Parker, 2006; Dower et al, 2004). Our above results indicate that in wild-type cells, GFP-RZ mRNA is indeed unstable, and that further destabilization of the mRNA by the presence of a PTC might have escaped detection. We therefore reasoned that destabilization of an unadenylated mRNA by NMD might be observed when the contribution of the cytoplasmic exosome to the decay of the mRNA is removed.
We measured decay rates of GFP(PTC)-RZ mRNA containing a PTC at codon 67 (Baker and Parker, 2006) in wild-type and ski7Δ cells. In contrast to the stabilization of GFP-RZ mRNA in ski7Δ cells, GFP(PTC)-RZ mRNA was unstable in both wild type and ski7Δ strains (Figure 1 E&F). Importantly, in ski7Δ cells GFP(PTC)-RZ mRNA stability was reduced compared to GFP-RZ mRNA (2 min versus 8 min; Figure 1 C&E), indicating that normal and premature translation termination events are distinguishable in the absence of a poly(A) tail.
Unadenylated mRNAs Harboring a PTC are Targets for NMD
One explanation for GFP(PTC)-RZ mRNA instability in ski7Δ cells is that it is degraded by the NMD pathway, while GFP-RZ mRNA is not. We therefore, tested if rapid degradation of GFP(PTC)-RZ mRNA required the NMD machinery. Specifically, we measured decay of GFP(PTC)-RZ mRNA in a mutant yeast strain lacking both Ski7p and an essential component of the NMD machinery, Upf1p (Leeds et al, 1991).
Analysis of GFP(PTC)-RZ RNA revealed significant stabilization of the mRNA in ski7Δupf1Δ cells (half-life=6.4 min; Figure 1G). In contrast, turnover of GFP-RZ mRNA was essentially unaffected by deletion of UPF1 (Figure 1 C&D). Thus, mRNA destabilization caused by the introduction of a PTC observed in ski7Δ cells was reversed by inactivation of the NMD pathway. The presence of an mRNA poly(A) tail is therefore not required for distinguishing a normal stop codon from a PTC by the NMD surveillance machinery.
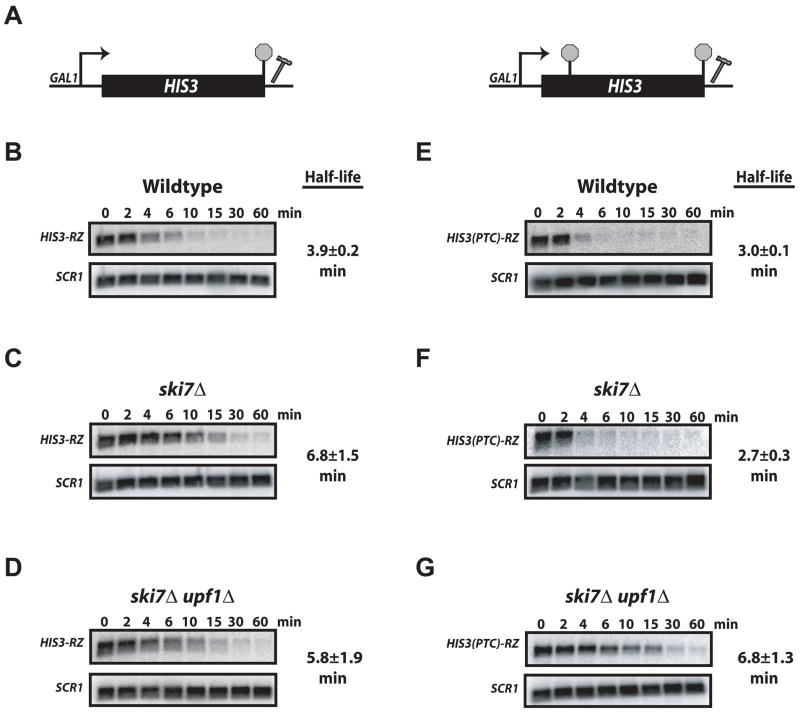
The experiments described above utilize the well characterized GFP-RZ reporter to evaluate decay of unadenylated mRNA (Baker and Parker, 2006; Dower et al, 2004). To ensure our results were not restricted to this heterologous reporter, we analyzed yeast HIS3 harboring a hammerhead ribozyme 55 nt downstream of the natural stop codon. As shown in Figure 2, HIS3-RZ mRNA in ski7Δ cells was destabilized by introduction of a PTC (compare C&F; Supplemental Figure 1), and rapid turnover of the PTC-containing mRNA required a functional NMD pathway (compare F&G). Thus, targeting of an unadenylated nonsense mRNA to NMD is not limited to the GFP reporter, and also can occur for yeast genes lacking a poly(A) tail. While detection of the destabilization of unadenylated mRNA by a PTC required that cytoplasmic exosome activity be eliminated, these results demonstrate that the NMD machinery is capable of substrate recognition independent of a poly(A) tail.
Figure 2. Unadenylated HIS3(PTC)-RZ mRNA is Targeted for NMD.
Stability of HIS3-RZ and HIS3(PTC)-RZ mRNA was determined as described in Figure 1.
Pab1p is Not Required for Destabilization of PTC-containing mRNA
Our findings with unadenylated mRNAs indicate that an mRNA poly(A) tail is not required for NMD in yeast. An association between PAB1 and an mRNA may, however, occur independently of the poly(A) tail. Indeed, interactions between PAB1 and eIF4G and eRF3 are well characterized (reviewed in Mangus et al, 2003). Therefore, to determine whether PAB1 itself is necessary for recognition of an mRNA by NMD, we evaluated levels of PGK1 reporter mRNA in yeast cells lacking Pab1p. Deletion of PAB1 is lethal, but inviability can be suppressed by a variety of secondary mutations, including, deletion of RRP6 (Dunn et al, 2005).
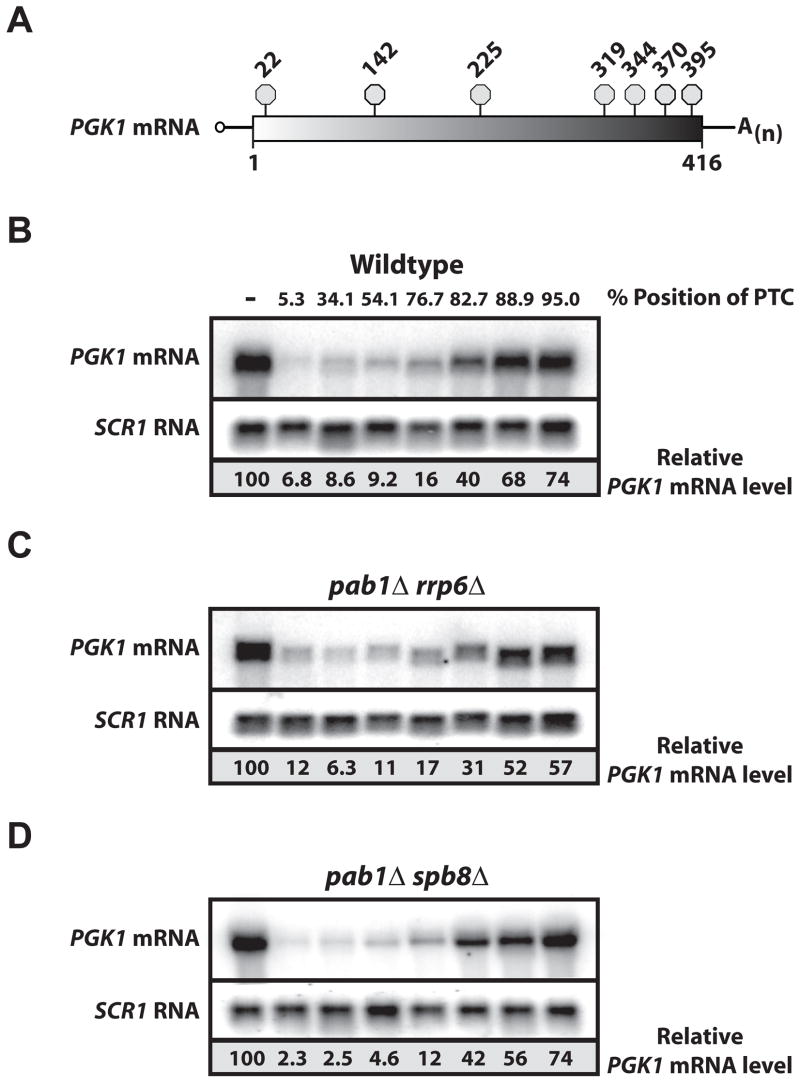
Levels of PGK1 reporter mRNA harboring a PTC at several positions within the ORF (Figure 3A) were evaluated in wild-type and pab1Δrrp6Δ cells. In wild-type, mRNAs harboring a nonsense codon were less abundant relative to wild-type PGK1 mRNA, and the decrease in nonsense mRNA was successively less dramatic as the PTC was positioned further 3′ within the PGK1 ORF, as previously reported (Figure 3B; Supplemental Figure 2; Cao and Parker, 2003; Peltz et al, 1993). Importantly, destabilization of the PTC-containing PGK1 mRNA in wild-type cells was dependent upon a functional NMD pathway (KEB and R Parker unpublished data). Analysis of reporter mRNAs in pab1Δrrp6Δ cells revealed the relative abundance of PGK1 mRNAs to be essentially identical to levels observed for wildtype (Figure 3C). Specifically, RNA levels were decreased for PGK1 mRNAs harboring PTCs, and the decrease correlated with the position of the PTC within PGK1. Our findings indicate that PTC-containing PGK1 mRNA is distinguished from wild-type mRNA in the absence of Pab1p.
Figure 3. Nonsense-containing PGK1 mRNA is Destabilized in the Absence of PAB1.
A. Schematic of reporter mRNA highlighting positions within the PGK1 ORF (box) of premature nonsense codons (stop signs). Northern blot analysis of steady-state PGK1 reporter mRNA in wildtype (B), pab1Δrrp6Δ (C) or pab1Δspb8Δ (D) cells. PTC location indicated as percent position into PGK1 ORF. RNA levels were normalized to SCR1 RNA and shown as percent level relative to PGK1 mRNA terminating at the natural stop codon for experiment shown; results for multiple analyses are shown in Supplemental Figure 2.
We ruled out an independent effect of the rrp6Δ allele on NMD through analysis of PGK1 reporter mRNA in pab1Δrrp6Δ cells complemented with PAB1. We demonstrate that destabilization of PTC-containing PGK1 mRNA was maintained in these cells (Supplemental Figure 3). We further analyzed NMD in a second yeast suppressor strain deleted for PAB1. In pab1Δspb8Δ cells (Boeck et al, 1998), nonsense and wild-type PGK1 mRNA levels resembled those observed in both wild-type and pab1Δrrp6Δ cells (Figure 3D). Thus, PTC-containing mRNAs are detected and destabilized in yeast cells in the absence of Pab1p.
Reduction of PTC-containing mRNA in the Absence of Pab1p Requires the NMD Pathway
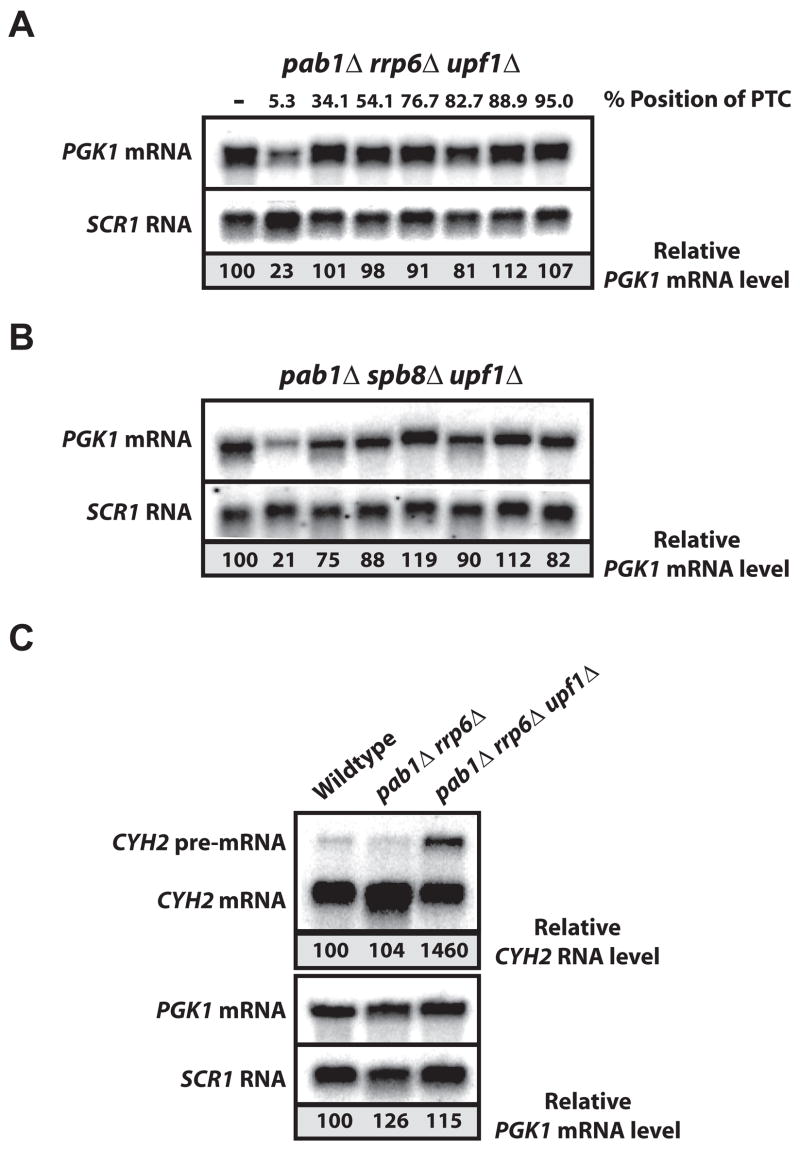
To determine if decreased RNA abundance of nonsense-containing PGK1 mRNAs in pab1Δ mutants was a consequence of NMD, we measured RNA levels in pab1Δrrp6Δ and pab1Δspb8Δ strains that also harbored a lesion in UPF1. In pab1Δrrp6Δupf1Δ and pab1Δspb8Δupf1Δ cells, nonsense PGK1 mRNA levels were similar to wild-type PGK1 mRNA (Figure 4 A&B; see Supplemental Material). These data demonstrate that the reduction in PTC-containing mRNA levels observed in the pab1Δ cells requires a functional NMD pathway and that substrates for NMD are efficiently recognized in the absence of Pab1p.
Figure 4. Degradation of Nonsense-containing mRNA in the Absence of Pab1p Requires a Functional NMD Pathway.
PGK1 reporter levels were determined by Northern blot analysis of RNA from pab1Δrrp6Δupf1Δ (A) and pab1Δspb8Δupf1Δ (B). RNA levels were normalized to SCR1 RNA loading control and shown as percent level relative to PGK1 mRNA void of a PTC for the experiment shown; results for multiple analyses are shown in Supplemental Figure 2. C. Northern blot analysis of endogenous CYH2 RNA and PGK1 reporter mRNA levels in wild-type, pab1Δrrp6Δ, and pab1Δrrp6Δupf1Δ cells. CYH2 pre-mRNA levels are normalized to CYH2 mRNA, while PGK1 mRNA levels are normalized to SCR1 RNA. Both are indicated as percent level relative to abundance in wild-type.
As a second measure of NMD in cells lacking PAB1, we analyzed levels of CYH2 pre-mRNA, an endogenous NMD target in yeast (He et al, 1993). We observed low levels of CYH2 pre-mRNA in wild-type and pab1Δrrp6Δ cells (Figure 4C). In contrast, CYH2 pre-mRNA in pab1Δrrp6Δupf1Δ mutants was elevated. We conclude that CYH2 pre-mRNA is targeted for NMD in the absence of Pab1p. Importantly, CYH2 mRNA and wild-type PGK1 mRNAs were similar despite the absence of Pab1p or a functional NMD pathway (Figure 4C). Therefore, in the absence of PAB1, the NMD machinery effectively discriminates between normal and nonsense-containing mRNAs.
DISCUSSION
Here we used ribozyme-cleaved reporter mRNA to evaluate the requirement of an mRNA poly(A) tail for NMD in yeast (Dower et al, 2004; Baker and Parker, 2006; Meaux and van Hoof, 2006). We show that unadenylated mRNA can be destabilized by introduction of a PTC, and that mRNA instability requires the NMD factor Upf1p (Figures 1&2). We further demonstrate that in yeast lacking the poly(A) tail binding protein, PAB1, mRNAs harboring a PTC are recognized and their levels reduced by the NMD machinery (Figures 3&4). Our findings show that the ability to discriminate between a normal and premature translation termination event can occur independently of an mRNA poly(A) tail or PAB1.
The faux UTR model for recognition of nonsense-containing mRNAs posits that translation termination at a PTC is intrinsically aberrant due to the failure of the terminating ribosome to engage in stabilizing interactions with the mRNA 3′ UTR (Amrani et al, 2006). According to the model, stabilizing signals are communicated to the ribosome by the proximal mRNP that includes the poly(A) tail and PAB1. An important tenet of this model is that a stop codon will be recognized as normal only if translation termination occurs sufficiently close to the poly(A) tail and its bound PAB1, and, therefore, in the absence of these elements, ribosomes terminating at a natural stop codon would fail to engage in the stabilizing interactions and be targeted for NMD. We find that unadenylated mRNA terminating at a natural stop codon is not stabilized by inactivation of the NMD pathway in either wild-type or ski7Δ cells (Baker and Parker, 2006; Figure 1&2, C versus D). Moreover, normal PGK1 and CYH2 mRNA are not targeted to NMD in the absence of PAB1 (Figure 4C). Therefore, transmission of a positive signal from PAB1 to the terminating ribosome is neither required for the stability of a ‘normal’ mRNA nor its ability to escape NMD.
Our findings are in conflict with interpretations of tethered function analysis used to support the faux UTR model. Tethering of PAB1 to mRNA downstream and proximal to a PTC is sufficient to stabilize an NMD substrate in both yeast and Drosophila (Amrani et al, 2004; Behm-Ansmant et al, 2007). Based on these findings, it has been interpreted that proximity of PAB1 to the PTC leads to redefinition of the premature termination event as normal and that PAB1 is critical for signaling proper translation termination. Our findings, however, demonstrate that PAB1 is dispensable for normal and aberrant translation termination to be distinguished, and necessitates reevaluation of this interpretation. Interestingly, mRNA stabilization by tethered PAB1 is not specific to NMD substrates and was first demonstrated for a ‘normal’ mRNA (Coller et al, 1998), illustrating that enhanced mRNA stability observed when PAB1 is tethered can occur independently of a role for PAB1 in NMD.
In Drosophila cells, nonsense-containing mRNA is stabilized by depletion of cytoplasmic PAB1 (PABPC1) by RNA interference (Behm-Ansmant et al, 2007). These data are in opposition to what is anticipated according to the faux UTR model (see above). These findings were, however, interpreted to indicate that PABPC1 was in some way required for NMD. The inconsistency between this conclusion and our findings may indicate a divergence in mechanism for NMD between flies and yeast. However, it is likely that depletion of PABPC1 impacts multiple steps in mRNA metabolism (e.g. 3′ end formation, nucleocytoplasmic transport, translation, stability) that might lead to stabilization of a PTC-containing mRNA indirectly. Saccharomyces cerevisiae strains suppressed for the essential function of PAB1 greatly facilitated analysis of PAB1 function in NMD by limiting these complications.
Our results establish that an interaction between the terminating ribosome and PAB1 does not determine whether a stop codon is perceived as normal or premature. Moreover, DSE elements previously defined as critical for NMD of PTC-containing PGK1 also appear immaterial in our studies, as PTCs downstream of the characterized elements in PGK1 mRNA still elicit NMD. While mRNAs with extended 3′ UTRs in yeast, Drosophila and mammals can be NMD substrates (Muhlrad and Parker, 1999, Behn-Ansmant et al, 2007; Buhler et al, 2006), not all mRNA with long 3′ UTRs are destabilized by the NMD pathway (Behn-Ansmant et al, 2007). The 6 nucleotides remaining downstream of the natural stop codon after cleavage of GFP-RZ mRNA further supports that NMD is not dependent on a 3′ UTR generated by conventional 3′ end formation (Figure 1; Baker and Parker, 2006). Finally, EJC complexes are not required for NMD in yeast, C. elegans, and Drosophila. Given the various second signals implicated in NMD substrate recognition, we predict that redundancy exists in the ability of aberrant mRNA to trigger NMD, although we can not exclude the possibility that additional elements necessary for NMD substrate discrimination remain to be uncovered. Future research evaluating the importance of coding region sequence and length, and features of the elongating polypeptide or ribosome itself in NMD are ongoing.
EXPERIMENTAL PROCEDURES
Strains
ski7Δ, upf1Δ, and ski7Δupf1Δ yeast are isogenic to wild-type BY4741 (Open Biosystems). The ski7Δupf1Δ mutant was constructed by crossing ski7Δ::HYG (Wilson et al, 2007) to upf1Δ::NEO and sporulating.
Wild-type, pab1Δrrp6Δ and pab1Δspb8Δ yeast strains have been described (Cao and Parker, 2003; Dunn et al, 2005). yKB300 (pab1Δrrp6Δupf1Δ [PAB1]) was generated by deleting UPF1 in pab1Δrrp6Δ cells complemented with pKB269 (PAB1+) by the method of Longtine et al (1998). yKB301 (pab1Δrrp6Δupf1Δ) was made by plating yKB300 on media containing 5′ fluoro-orotic acid to select for loss of PAB1+URA3+ plasmid. Loss of the PAB1 plasmid was confirmed by Western blotting. pab1Δspb8Δupf1Δ (yKB310) cells were generated by deleting UPF1 (Longtine et al, 1998).
Plasmids
GFP-RZ plasmid pAV298 contains 1.3 kbp SpeI and XhoI fragment of GAL-GFP-RZ (Dower et al, 2004) in pRS416 (Sikorski and Hieter, 1989). GFP(PTC)-RZ plasmid pAV339 is similarly derived from pKB214 (Baker and Parker, 2006). HIS3-RZ has been described (Meaux and van Hoof, 2006). To generate HIS3(PTC)-RZ (pAV462), the sequence 5′ T ATC TAA GAA TTC TAA CA (PTC in bold) was inserted in the NdeI site of HIS3-RZ.
Plasmids for wild-type PGK1 (pRP469) and PTCs at codon positions 22, 142, 225, 319 have been described (Cao and Parker, 2003), while PTCs at codon positions 344, 370 and 395 were created by site-directed mutagenesis of pRP469 (KEB and R Parker, manuscript in prep).
RNA Analysis
Steady state RNA levels were measured from cells grown in SC-URA+ 2% galactose/1% sucrose at 24 °C and harvested at OD600 = 0.4–0.5. For RNA decay measurements, yeast strains harboring GFP-RZ reporter plasmids were grown at 30 °C in SC-URA + 2% galactose to mid log phase. Media was replaced with SC-URA + 4% glucose and culture aliquots removed over time. RNA was extracted and blotted by standard methods. Northern blots were probed with 32P 5′ radio-labeled oligonucleotides specific for GFP (5′GCTGTTACAAACTCAAGAAGGACCATGTGG 3′), HIS3 (5′ CTACCACCGCTCTGGAAAGTGCCTCATCCA 3′), PGK1 (oRP121; Decker and Parker, 1993), CYH2 (5′ CCATACCTCTACCACCGGGGTGCTTTCTGTGCTTACCG), or the RNA of the signal recognition particle (SCR1 RNA; 5′GTCTAGCCGCGAGGAAGG 3′). RNA levels were quantified using a STORM phosphoimager (GE Healthcare).
Supplementary Material
Acknowledgments
GAL1-GFP-RZ plasmid was provided by Michael Rosbash (Brandeis Univ., MA), polyclonal antibodies to ribosomal protein L5 by John Woolford (Carnegie Mellon Univ., PA) and pab1Δrrp6Δ and pab1Δspb8Δ yeast by Charles Cole (Dartmouth Medical School, NH). We thank Tim Nilsen, Jo Ann Wise, Jonatha Gott, Jeff Coller and members of the Coller and van Hoof laboratories for critical reading of the manuscript. Funding was provided from the Pew Scholarship program in the Biomedical Sciences and NIH (GM069900) to AvH, and by Case Western Reserve University to KEB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol. 2006;7:415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- Baker KE, Parker R. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol. 2004;16:293–299. doi: 10.1016/j.ceb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Baker KE, Parker R. Conventional 3′ end formation is not required for NMD substrate recognition in Saccharomyces cerevisiae. RNA. 2006;12:1441–1445. doi: 10.1261/rna.92706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck R, Lapeyre B, Brown CE, Sachs AB. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Steiner S, Mohn F, Paillusson A, Muhlemann O. EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′. UTR length Nat Struct Mol Biol. 2006;13:462–464. doi: 10.1038/nsmb1081. [DOI] [PubMed] [Google Scholar]
- Cao D, Parker R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell. 2003;113:533–545. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- Coller JM, Gray NK, Wickens MP. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson MR, Leeds PF. Looking at mRNA decay pathways through the window of molecular evolution. Curr Opin Genet Dev. 2003;13:207–214. doi: 10.1016/s0959-437x(03)00014-5. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Dower K, Kuperwasser N, Merrikh H, Rosbash M. A synthetic A tail rescues yeast nuclear accumulation of a ribozyme-terminated transcript. RNA. 2004;10:1888–1899. doi: 10.1261/rna.7166704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EF, Hammell CM, Hodge CA, Cole CN. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1: connect mRNA biogenesis to export. Genes Dev. 2005;19:90–103. doi: 10.1101/gad.1267005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez CI, Ruiz-Echevarria MJ, Vasudevan S, Henry MF, Peltz SW. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol Cell. 2000;5:489–499. doi: 10.1016/s1097-2765(00)80443-8. [DOI] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- He F, Peltz SW, Donahue JL, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1-mutant. Proc Natl Acad Sci U S A. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmiak HA, Maquat LE. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med. 2006;12:306–316. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D, Plasterk RH, Johnstone IL, Caceres JF. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 2007;21:1075–1085. doi: 10.1101/gad.417707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Meaux S, van Hoof A. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA. 2006;12:1323–1337. doi: 10.1261/rna.46306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay: is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA. 1999;5:1299–1307. doi: 10.1017/s1355838299990829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira CC, McCarthy JE. The relationship between eukaryotic translation and mRNA stability. A short upstream open reading frame strongly inhibits translational initiation and greatly accelerates mRNA degradation in the yeast Saccharomyces cerevisiae. J Biol Chem. 1995;270:8936–8943. doi: 10.1074/jbc.270.15.8936. [DOI] [PubMed] [Google Scholar]
- Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Lucy Raymond F, Nguyen L, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Green PJ. Premature nonsense codons decrease the stability of phytohemagglutinin mRNA in a position-dependent manner. Plant J. 1996;10:415–424. doi: 10.1046/j.1365-313x.1996.10030415.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Gudikote JP, Olivas OR, Wilkinson MF. Boundary-independent polar nonsense-mediated decay. EMBO Rep. 2002;3:274–279. doi: 10.1093/embo-reports/kvf036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil JE, Beemon KL. A 3′ UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA. 2006;12:102–110. doi: 10.1261/rna.2129806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch EM, Jacobson A. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 1999;18:6134–6145. doi: 10.1093/emboj/18.21.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Meaux S, van Hoof A. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics. 2007 doi: 10.1534/genetics.107.073205. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, LaDuca JP, Maquat LE. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998a;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun X, Qian Y, Maquat LE. Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998b;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.