Abstract
The use of “anchor-fixed” altered peptide ligands is of considerable interest in the development of therapeutic vaccines for cancer and infectious diseases, but the mechanism by which successful altered peptide ligands elicit enhanced immunity is unclear. In this study, we have determined the crystallographic structure of a major tumor rejection Ag, gp100209–217, in complex with the HLA-A*0201 (HLA-A2) molecule, as well as the structure of a modified version of the peptide which substitutes methionine for threonine at position 2 (T2M; gp100209–2M). The T2M-modified peptide, which is more immunogenic in vitro and in vivo, binds HLA-A2 with a ∼9-fold greater affinity and has a ∼7-fold slower dissociation rate at physiological temperature. Within the limit of the crystallographic data, the T2M substitution does not alter the structure of the peptide/HLA-A2 complex. Consistent with this finding, in peripheral blood from 95 human subjects, we were unable to identify higher frequencies of T cells specific for either the native or modified peptide. These data strongly support the conclusion that the greater immunogenicity of the gp100209–2M peptide is due to the enhanced stability of the peptide/MHC complex, validating the anchor-fixing approach for generating therapeutic vaccine candidates. Thermodynamic data suggest that the enhanced stability of the T2M-modified peptide/HLA-A2 complex is attributable to the increased hydrophobicity of the modified peptide, but the gain due to hydrophobicity is offset considerably by the loss of a hydrogen bond made by the native peptide to the HLA-A2 molecule. Our findings have broad implications for the optimization of current vaccine-design strategies.
The development of therapeutic vaccines based on tumor-associated Ags (TAAs)3 presented by class I MHC molecules is one of the goals of cancer immunotherapy. As most TAA are self-Ags (1), the Ag-specific CTL repertoires may be significantly reduced during the processes of negative selection, leaving a T cell repertoire that is poorly effective at mounting a productive antitumor immune response. Efforts to use unmodified forms of peptide immunogens to elicit antitumor responses in vivo have been largely unsuccessful, leading investigators to modify naturally occurring tumor Ags. Indeed, a number of altered peptide ligands (APL) capable of eliciting enhanced immunity to tumor-associated epitopes have been developed (e.g., Refs. 2-4). The majority of these APL are “anchor-fixed” variants, in which suboptimal primary anchors have been replaced with more optimal amino acids.
The mechanisms underlying the enhanced immunogenicity of these APL have remained incompletely elucidated. Speculations on how APL elicit enhanced immune responses have included the possibility that APL present a different peptide conformation to T cells, thus stimulating a different set of TCRs which are capable of cross-reacting with the native epitope. Indeed, anchor-fixing does not always result in enhanced immunogenicity (e.g., Ref. 5), and in some cases this has been shown to result from structural alterations (6, 7). TCRs can be exquisitely sensitive to changes in the structure of the peptide, in some cases sensing alterations resulting from removal or addition of a single methylene group (6, 8).
Others have suggested that native TAA function as partial agonists or antagonists, stimulating anergy in responding cells, whereas APL trigger T cells to signal in a more productive fashion (9). A third hypothesis is that the CTL repertoire specific for TAA remains intact and inducible, but weak peptide affinities or fast peptide dissociation rates preclude the formation of a stable immunological synapse, preventing sufficient signaling through the TCR to convert naive precursor TAA-specific CTLs into differentiated CTLs capable of lysing TAA-presenting tumor cells (10).
In this study, we focus on the 209–217 epitope of the gp100 membrane glycoprotein (gp100209; sequence ITDQVPFSV), for which the anchor-fixing substitution of the threonine at position 2 with methionine has previously been described (5). The modified peptide (IMDQVPFSV; gp100209–2M) is considerably more immunogenic in vitro and in vivo (5, 11), and its use in clinical trials is correlated with a higher incidence of objective antitumor immunity (2, 12). Using a human-mouse chimera in which the gp100209 epitope was knocked out, we recently found that absence of the target epitope did not substantially increase the ability of the native peptide to immunize, whereas the T2M modification to gp100209 dramatically increased its immunogenicity (10), demonstrating that the poor immunogenicity of the native peptide is not due to negative selection of gp100209-specific T cells.
Initial studies of the enhanced immuogenicity of the gp100209–2M peptide focused on its ∼9-fold tighter steady-state binding affinity to the restricting MHC molecule (HLA-A*0201) (5). We recently reported that perhaps a more important correlate with the increased immunogenicity of the gp100209–2M peptide is its decreased dissociation rate from HLA-A*0201 (HLA-A2), or equivalently, the greater half-life of the HLA-A2 complex with the modified peptide (10). At physiological temperature, the half-life of the gp100209–2M/HLA-A2 complex is ∼7-fold greater than the half-life of the complex with the native peptide. Consistent with this measurement, in vitro, T cell recognition of APCs pulsed with the native gp100209 peptide is substantially reduced when a 24-h time delay is introduced into the experiment, whereas those pulsed with the T2M-modified peptide are unaffected (10).
To further investigate the effects of the gp100209 T2M substitution, in this study we determined the crystallographic structures of the native and modified gp100209 peptides bound to HLA-A2. The data indicate that the structures are indistinguishable. Consistent with the structural data, in peripheral blood samples from 95 patients with metastastic melanoma, we were unable to detect different frequencies of T cells specific for the native or T2M modified peptides. These findings strongly support the interpretation that the greater immunogenicity of the T2M-modified peptide is due to stabilization of the peptide/MHC complex, rather than activation of subsets of high-avidity, cross-reactive T cells. These data provide an important validation for anchor-fixing as a viable strategy for generating APLs with improved immunogenicity.
Measurements of peptide dissociation thermodynamics suggest that the enhanced stability of the complex with the T2M-modified peptide is attributable to its increased hydrophobicity. However, the structure of the native gp100209/HLA-A2 complex shows that the threonine at position 2 forms a hydrogen bond to Glu63 of the HLA-A2 α1 helix. The thermodynamic measurements suggest that loss of this hydrogen bond with the T2M-modified peptide considerably offsets the effects of increased peptide hydrophobicity. Coupled with the crystallographic structures, these data suggest a route for making additional modifications to the gp100209–217 peptide that result in even greater peptide/MHC stability, with possible concomitant increases in immunogenicity. Such peptide modifications may have general applicability beyond the gp100209 tumor-associated Ag.
Materials and Methods
Proteins and peptides
Protein was produced by the standard technique of refolding bacterially expressed HLA-A2 (truncated at the transmembrane region) and β2-microglobulin (β2m) inclusion bodies in the presence of excess peptide (13). Refolded protein was purified via ion-exchange and size-exclusion chromatography. Peptides were synthesized and purified to >95% commercially (SynPep). Extinction coefficients at 280 nM (units of M−1 cm−1) were 93439 for peptide/HLA-A2 complexes with unlabeled peptide and 103200 for complexes with labeled peptide.
X-ray crystallography
Peptide/HLA-A2 crystals were grown from 25 mM MES, pH 6.5, 24% polyethylene glycol 3350 using sitting drop/vapor diffusion. For cryoprotection, crystals were transferred to 30% polyethylene glycol 3350, 20% glycerol. Diffraction data were collected at the Argonne Structural Biology Center using the 19BM beamline. Data reduction was performed with HKL2000 (14). Structures were solved using molecular replacement with MOLREP from CCP4 (15). The search model was Protein Data Bank entry 1B0R (16), using only the coordinates for the H chain and β2m. Rigid body refinement, TLS refinement, and restrained refinement were performed with Refmac5 (17). Anisotropic and bulk solvent corrections were performed in all steps of refinement. TLS groups were chosen for residues 1–182 and 183–275 of the H chain, 0–99 of β2m, and 1–9 of the peptides. After TLS refinement, it was possible to unambiguously position the peptides in 2Fo-Fc maps. Waters were added using ARP/wARP (18). Graphical evaluation of the model and fitting to maps was performed using XtalView (19). Procheck (20) and the template structure check in the WHATIF server (21) were used to evaluate the quality of the structure during and after refinement. Structure factors and coordinates have been submitted to the Protein Data Bank (entries 1TVB and 1TVH). Cavity calculations were performed with SURFNET (22).
IFN-γ ELISPOT assays
PBMC from 95 HLA-A*0201+ patients at high risk for recurrence of melanoma were collected using apheresis and cryopreserved during their course of immunization with the gp100209–2M peptide in protocols reviewed and approved by the Institutional Review Board of the National Institutes of Health. All patients signed an Institutional Review Board-approved consent form and had histologically confirmed melanoma. A total of 106 PBMC samples from these patients were assayed for peptide-specific IFN-γ ELISPOT. The PBMC were collected at various times during the course of immunization, but assays of these samples with the native and modified peptides were always done at the same time. Cryopreserved PBMC were thawed and rested in culture medium overnight and then were added to 1 μM peptide-pulsed C1R-A2 target cells at a 1:1 ratio (105 of each effector and target cells) in triplicates of 96-well ELISPOT plates (precoated with anti-human IFN-γ Abs from BioSource). After a 24-h incubation, the plates were washed, coated with biotinylated anti-IFN-γ Abs (BD Pharmingen), followed by alkaline phosphatase-conjugated avidin incubation. The spots were developed with alkaline phosphatase substrate. Numbers of spots were corrected by subtracting background spots caused by PBMC incubation with C1R-A2 cells pulsed with an irrelevant control peptide (gp100280; YLEPGPVTA). Nonspecific spots were between 0 and 14/105 cells.
Peptide dissociation kinetics
Dissociation kinetics were measured using fluorescence anisotropy with a Beacon 2000 polarization instrument (Invitrogen Life Technologies) as previously described (23). Briefly, 7.5 nM peptide/HLA-A2 loaded with a fluorescently labeled peptide was mixed with 7.5 μM unlabeled peptide, and the decrease in anisotropy was measured as a function of time. Data were fit to single or biphasic functions of the form
where y0 is the baseline offset, the summation is over the number of phases i (1 or 2), Ai is the amplitude for phase i, ki is the rate constant for phase i, and t is the time. Data were analyzed using the program Origin (OriginLab). Error analysis was performed using standard error propagation techniques (24). All measurements were performed in 10 mM HEPES, 150 mM NaCl, pH 7.4 or 6.5, as indicated.
Results
The crystal structures of the gp100209 and gp100209–2M peptides bound to HLA-A2 are indistinguishable
The crystal structures of the native gp100209 and anchor-modified gp100209–2M peptides bound to HLA-A2 were determined at 1.8 Å resolution via molecular replacement. Both complexes crystallized in space group P21 with two molecules per asymmetric unit (molecules “A” and “B”). For both complexes, unambiguous electron density was observed for the peptides (Fig. 1). Crystallographic data and refinement statistics are provided in Table I.
FIGURE 1.
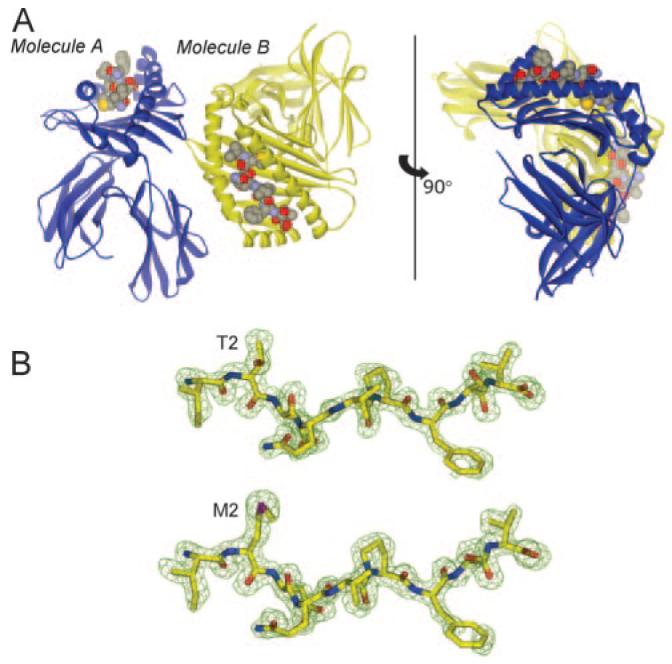
Overview of the gp100209/HLA-A2 and gp100209–2M/HLA-A2 crystal structures. A, Orientation of the two molecules observed in the two asymmetric units. Molecule A is blue and molecule B is yellow. This image shows the T2M-modified structure, but the overall structure with the native peptide is identical. B, 2Fo-Fc electron density maps contoured at 1 σ for the peptides from the A molecules. Native peptide is on top, the T2M variant is on the bottom.
Table I.
X-ray data and refinement statistics
| Complex | gp100209/HLA-A2 | gp100209–2M/HLA-A2 |
|---|---|---|
| Source | APS 19BM | APS 19BM |
| Space group | P21 | P21 |
| a (Å) | 58.39 | 58.41 |
| b (Å) | 84.45 | 84.49 |
| c (Å) | 84.02 | 84.01 |
| β | 90.01° | 90.11° |
| Molecules/a.u. | 2 | 2 |
| Resolution, Å | 20–1.8 | 20–1.8 |
| Total number of reflections | 74684 | 74189 |
| Mosaicity | 0.36° | 0.60° |
| Completeness, % | 99 (98.2)a | 98.7 (97.0) |
| I/σ | 16.8 (3.7) | 16.3 (2.7) |
| Rmerge, % | 8.5 (34) | 6.5 (40) |
| Average redundancy | 3.9 (3.6) | 2.8 (2.6) |
| Rwork, % | 16.5 | 18.5 |
| Rfree, % (no. of reflections) | 21.6 (3748) | 24.7 (3703) |
| Average B factor, Å2 | 12.7 | 14.5 |
| Ramachandran plot | ||
| Most favored, % | 92.0 | 91.9 |
| Allowed, % | 7.7 | 7.8 |
| Generously allowed, % | 0.3 | 0.3 |
| Disallowed, % | 0.0 | 0.0 |
| RMS deviations from ideality | ||
| Bonds, Å | 0.018 | 0.022 |
| Angles | 1.946° | 2.140° |
| Coordinate error, Å | 0.08b | 0.11 |
Numbers in parenthesis refer to the highest resolution shell.
Mean estimate based on maximum likelihood methods.
In both the native and T2M-modified gp100209/HLA-A2 crystal structures, both molecules in the asymmetric unit are nearly identical. There are, however, differences in the positioning of Gln4, Phe7, and Ser8 of the peptide. As shown in Fig. 2, A and B, though, these differences are common to both the native and T2M-modified structures. Comparison of the two sets of peptides thus reveals that the T2M substitution has little, if any, effect on peptide conformation. For the A molecules, all common atoms in the peptides superimpose to a root mean square deviation (RMSD) of 0.20 Å. For the B molecules, the superimposition is 0.21 Å (a detailed comparison of the two structures is provided in Table II). The largest differences in the peptides are the side chains of positions 4, 7, and 8, the same positions which show differences between the A and B molecules (Fig. 3). Thus we cannot attribute the small differences between the native and modified peptides to the T2M substitution, as the differences are in the same positions and are of lower magnitude than the differences observed between the A and B molecules in both structures. The temperature factors for the atoms of these side chains do not differ significantly from the average in either structure. Thus, these differences may reflect different side chain conformations that interconvert on a slow timescale, captured by crystallization, or alternatively, the influence crystal packing has on otherwise mobile side chains. We note that similar differences in exposed side chains are seen elsewhere in the structures, for example Lys19 in β2m, a full 42 Å away from the site of the T2M substitution. The side chain differences are thus reminiscent of the kind of differences one observes when comparing different structures of the same protein. For example, when comparing the original structure of the Tax peptide bound to HLA-A2 reported in 1993 (25) with a higher resolution structure reported in 2000 (26), all atoms of the peptides superimpose to an RMSD of 0.21 Å, identical to the values obtained for the superimposition of the gp100209 and gp100209–2M peptides reported in this study (Table II).
FIGURE 2.
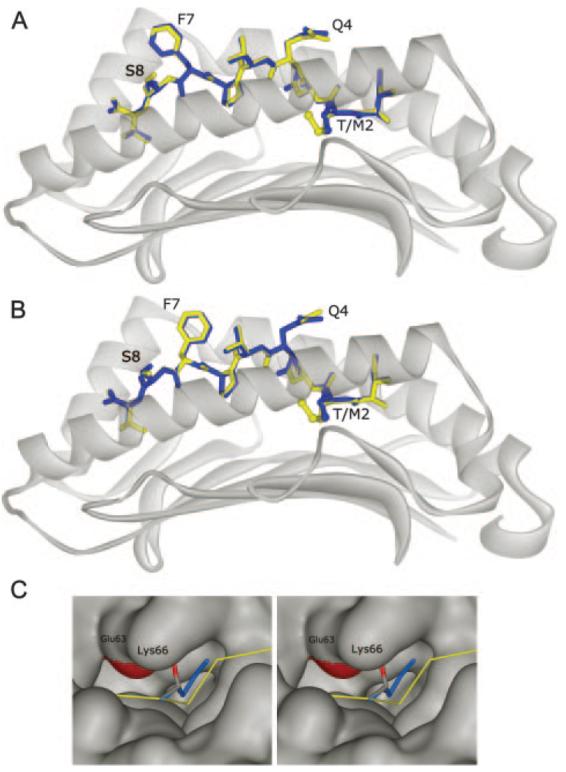
Superimposition of the peptides from the gp100209/HLA-A2 and the gp100209–2M/HLA-A2 crystal structures. The native peptide is blue, the T2M-modified peptide is yellow. A and B, Superimposition of the two A molecules (top) and B molecules (bottom). The P2 side chain is indicated as T/M2. Note the repositioning of Q4, F7, and S8 between the A and B molecules (compare top to bottom), but note also how these differences are the same whether one examines the structure with the native or T2M-modified peptide (compare blue to yellow in both images). C, Cross-eyed stereo image of the fit of the Thr2 and Met2 side chains in the HLA-A2 P2 pocket. Methionine is blue; the carbons of threonine are gray; and the oxygen is red. The surface of HLA-A2 is gray, except for the carboxylate of Glu63 which is red. Glu63 and Lys66 are indicated.
Table II.
Differences between the native and T2M-modified peptide/MHC structures are smaller than the differences between the two molecules in the asymmetric unitsa
| Comparison of the Two Molecules in the Asymmetric Units |
Comparison of Native and T2M- Modified Structures |
|||
|---|---|---|---|---|
| Region Compared | Native A onto native B |
T2M-modified A onto T2M-modified B |
Native A onto T2M-modified A |
Native B onto T2M-modified B |
| Whole complex | ||||
| All common atoms | 0.76 | 0.76 | 0.66 | 0.66 |
| Backbone | 0.35 | 0.29 | 0.15 | 0.30 |
| Peptide binding domain | ||||
| All atoms | 0.80 | 0.78 | 0.67 | 0.65 |
| Backbone | 0.46 | 0.33 | 0.18 | 0.41 |
| Peptide | ||||
| All common atoms | 1.10 | 1.05 | 0.20 | 0.21 |
| Backbone | 0.10 | 0.08 | 0.09 | 0.08 |
RMSD in angstroms.
FIGURE 3.
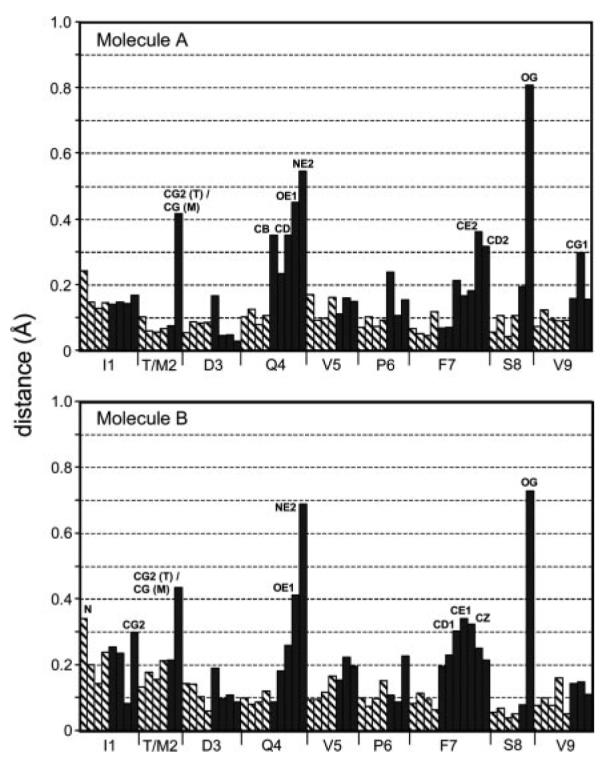
Interatomic distances between superimposed gp100209 and gp100209–2M peptides. Hatched bars indicate backbone atoms, dark gray indicate side chains. The figure corresponds to the superimpositions in Fig. 2, A and B. Note that the backbone differences are miniscule and without any trends, and that, excluding the site of the substitution, the most significant differences are for atoms in positions 4, 7, and 8, the same positions which show differences between the two molecules in each asymmetric unit. Atoms with distances ≥0.30 Å are labeled.
Comparing the backbones of the gp100209 and gp100209–2M peptides in the two structures, for both the A and B molecules, the backbone of the gp100209 peptide superimposes on that of the gp100209–2M peptide with an RMSD of ∼0.1 Å, near the estimated coordinate error (Table I). Thus, within the limit of the x-ray data, the T2M substitution has no effect on the structure of the peptide in the HLA-A2 binding groove.
Comparison of the HLA-A2 α1 and α2 helices in the two structures indicates that the T2M substitution does not induce changes into the structure of the peptide binding domain. The backbones of these helices superimpose to RMSDs of 0.09 and 0.11 Å, respectively. The side chains of HLA-A2 amino acids that contact TCRs in known TCR/peptide/HLA-A2 structures likewise show no significant differences. For example, the side chains of Lys66 and Gln155, implicated as important positions for most HLA-A2 restricted TCRs (27, 28), are virtually indistinguishable, with equivalent atoms differing by no more than 0.45 Å when the peptide binding grooves are superimposed. As with the peptides, the positions of some side chains differ slightly when comparing the A and B molecules. Again though, as summarized in Table II, these differences tend to be equal to or greater than any differences observed between the structures with the native and modified peptides.
The structure with the native peptide reveals how the polar side chain of Thr2 is accommodated in the apolar HLA-A2 P2 pocket, inserting itself into a network of hydrogen bonds strongly conserved in peptide/HLA-A2 structures (7, 16, 25, 26, 29-35). As shown in Fig. 4, the threonine at position 2 hydrogen bonds with Glu63 of the HLA-A2 H chain. Glu63 also hydrogen bonds with the conserved water molecule near the peptide amino terminus and forms a salt-bridge with Lys66. Lys66 in turn hydrogen bonds to the carbonyl oxygen of Thr2. Substitution of Thr2 with methionine removes the hydrogen bond between the side chain and Glu63, but Glu63, Lys66, and the position 2 carbonyl oxygen are undisturbed.
FIGURE 4.
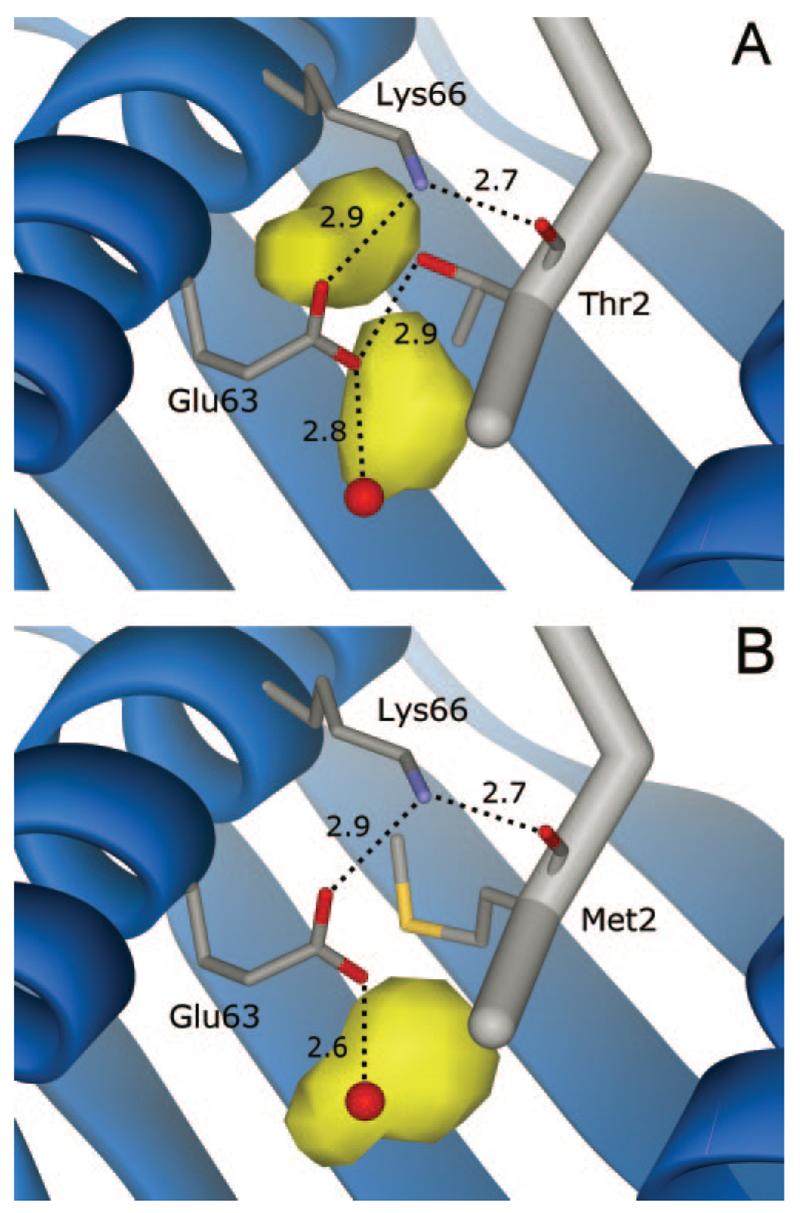
Cavities and hydrogen bonding in and around the P2 pocket in the two peptide/MHC structures. In the structure with the native peptide (A), the hydroxyl group of the Thr2 side chain hydrogen bonds with Glu63, a participant in a network of hydrogen bonds strongly conserved in peptide/HLA-A2 structures. B, The hydrogen bond to Glu63 is lost when Thr2 is substituted with Met. Numbers indicate hydrogen bond distances. The red sphere is a conserved water whose position is slightly altered in the T2M structure. Two large cavities are present within the P2 pocket when Thr is at position 2 (yellow polygons). The smaller cavity is occupied by the -S-CH3 moiety of the Met side chain in the modified structure. The larger cavity increases in response to removal of the Thr hydroxyl and repositioning of the γ carbon. This figure was generated using the A molecules, but the conclusions are identical if the B molecules are compared. A conserved hydrogen bond between Glu63 and the amide nitrogen of P2 is not shown for clarity.
As the two structures are so similar, one expectation is that replacement of the smaller threonine with the larger methionine should alter packing in and around the HLA-A2 P2 pocket. In the structure with the native peptide, cavity calculations (22) between the side chain of Thr2 and HLA-A2 reveal the presence of cavities in and around the P2 pocket, shown in Fig. 4A. The same calculation on the modified structure confirms that substitution of Thr2 with Met reduces total cavity volume. The larger cavity shown in Fig. 4A increases in volume from 15.4 to 25.6 Å3 in response to removal of the threonine hydroxyl and repositioning of the γ carbon (Fig. 4B). However, the smaller cavity (volume of 14.8 Å3) is not present in the modified structure, as this space is occupied by the -S-CH3 moiety of the methionine side chain. The total reduction in cavity volume upon making the T2M substitution is thus 4.6 Å3. The H chain atoms defining the cavities shown in Fig. 4 superimpose to an RMSD of 0.14 Å, indicating that the overall size and shape of the P2 pocket is not altered due to the T2M substitution.
It is informative to compare our findings with other structural studies of native and modified peptides presented by MHC molecules. In studying the Hb peptide presented by the murine class II MHC I-Ek, Kersh et al. (6) found that changing the P6 anchor from glutamic to aspartic acid resulted in a subtle (0.4–0.6 Å) but crystallographically significant shift in the position of the peptide backbone (6). Although the change was maximal at the site of the substitution, the effects propagated from the site of the substitution all the way to the peptide C terminus. No such trends are observed in our data; indeed, the backbone differences tend to be at or below the estimated coordinated error. Sharma et al. (7) obtained similar results with variants of the HER-2/neu peptide bound to HLA-A2: with the leucine at position 9 changed to valine, a significant change in the conformation of the peptide backbone in the center of the peptide was observed (7). Again, no such changes are detected in this study.
Other structural studies have revealed conformational changes in peptide/MHC complexes when the peptides are modified (e.g., Refs. 36 and 37), but our findings of no conformational changes occurring upon modifying the gp100209 peptide are not without precedent. With the RT309–317 peptide bound to HLA-A2, Kirksey et al. (32) found that changing the P1 residue from isoleucine to either phenylalanine or tyrosine did not alter the structure of the peptide/HLA-A2 complex (32), despite large changes in peptide/HLA-A2 stability and immunogenicity. Likewise, with the NY-ESO157–165 peptide bound to HLA-A2, modifications to P9 did not alter the structure (38). Finally, in a ternary complex between the A6 TCR and the Tax peptide presented by HLA-A2, the conformation of the Tax peptide was found to be identical whether a proline or an alanine was present at position 6, despite the fact that the P6A substitution changes the peptide from a strong agonist to an antagonist (8, 39).
T cells from vaccinated patients are unable to distinguish between the native and T2M-modified peptides
To investigate whether T cells can distinguish between the native and T2M-modified gp100209 peptides, 106 PBMC samples from 95 patients at high risk for recurrence of melanoma and who had been vaccinated with the T2M-modified peptide were assayed for peptide-specific IFN-γ release using ELISPOT. As shown in Fig. 5, across this large sample size, the number of cells activated by the native peptide is highly correlated with the number of cells activated by the T2M-modified peptide. The slope of the regression line, 1.1 ± 0.1, indicates that there is little, if any, trend indicating the existence of T cells that can discriminate between the two peptides. The slightly positive value of the slope in favor of the T2M-modified peptide may reflect its improved binding affinity and slower dissociation rate (5, 10).
FIGURE 5.
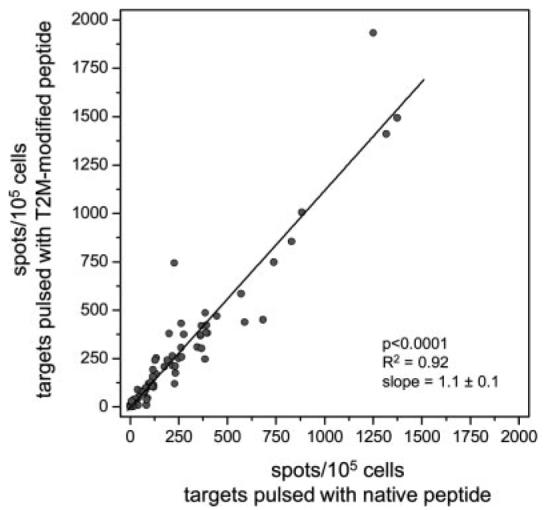
The frequency of T cells recognizing the native gp100209 peptide mirrors the frequency of those recognizing the T2M-modified peptide. The figure shows results from ELISPOT assays conducted on 106 PBMC from 95 individuals, identifying the number of Ag-specific T cells per 105 cells. The line represents a linear regression through the data, characterized by a slope of 1.1 ± 0.1.
Because ELISPOT only measures the frequencies of Ag-specific T cells, we could not confirm whether a gp100209–2M-reactive T cell was indeed gp100209-reactive. However, if some or all of the T cell subsets tested were capable of specifically recognizing only the T2M peptide, against which these patients were immunized, in Fig. 5 we would expect to see a large number of points “pinned” to the y-axis, with greatly diminished recognition of the native peptide. However, in 106 samples from 95 patients, the majority had nearly identical frequencies of gp100209 and gp100209–2M-specific T cells. This strongly suggests that T cells specific for gp100209 presented by HLA-A2 cannot distinguish between the native and T2M-modified peptide, in accordance with the crystallographic data. Very recent data indicate that the T cell reactivity against these peptides seen in ELISPOT is also a measure of their reactivity toward tumors, which likely present much lower amounts of the native peptide than is achieved with the 1 μM pulse to cultured APCs performed in this study (N. P. Restifo, manuscript in preparation).
Activation thermodynamics for dissociation indicate hydrophobicity as the mechanism for the improved stability of the gp100209–2M/HLA-A2 complex
The crystallographic structures described above do not directly indicate the physical mechanism behind the increased stability of the gp100209–2M/HLA-A2 complex. Filling cavity space is an established way of enhancing protein stability, but at 24–33 cal/mol/Å3 of cavity space (40), the net decrease in volume of only 4.6 Å3 (Fig. 4) provides for very little gain.
To gain insight into how substitution of Thr2 with Met enhances peptide/HLA-A2 stability, we examined peptide dissociation kinetics as a function of temperature, determining activation thermodynamics for peptide dissociation via transition state analysis. Peptide dissociation measurements were performed with soluble, purified peptide/HLA-A2 molecules using steady-state fluorescence anisotropy. We used fluorescent derivatives of the gp100209–217 and gp100209–2M peptides with a single amino acid substituted with a fluorescein-conjugated lysine. Data were collected over the range of 4–37°C in the presence of a large excess of unlabeled peptide. Fig. 6 shows dissociation data for peptides modified at Val5. The decreased dissociation rate for the T2M-modified peptide is evident at all temperatures studied.
FIGURE 6.
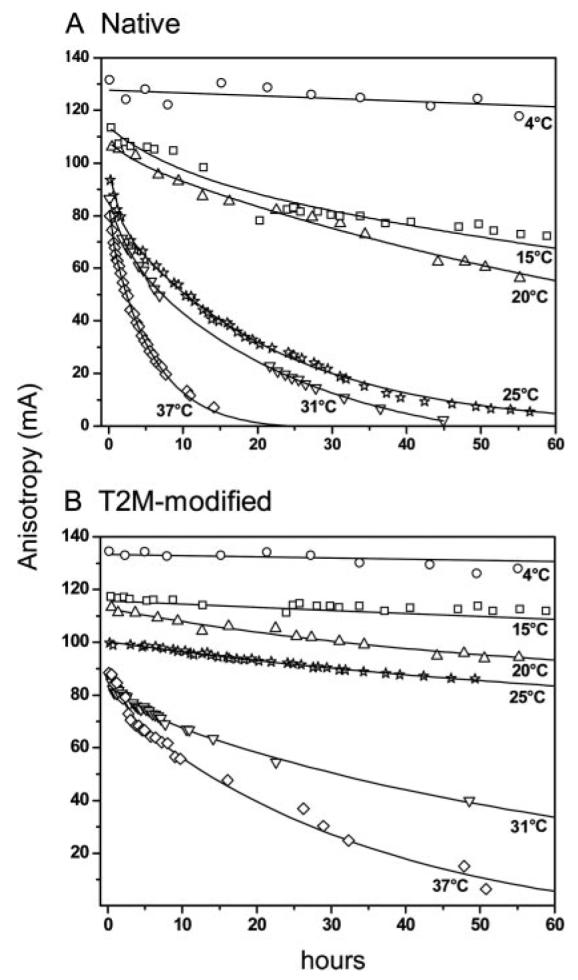
Dissociation of the gp100209 (A) and gp100209–2M (B) peptides from HLA-A2 as a function of temperature. The decreased dissociation rate of the T2M-modified peptide is evident at all temperatures. Data for lower temperatures were taken for hundreds of hours (>2000 for 4°C), but are not shown for clarity. Lines represent exponential fits to the data as described in the Materials and Methods.
Consistent with similar quantitative analyses of peptide dissociation from class I MHC molecules (23, 41-43), the majority of the data in Fig. 6 were best fit with a double-exponential decay function, consisting of a slow phase accounting for the bulk of the data and a minor, faster phase typically accounting for much smaller (5–10%) of the total amplitude. In similar measurements, the minor phase has been attributed to the presence of a small amount of peptide/H chain heterodimer, from which peptide more rapidly dissociates (23, 41). The major phase thus represents the more physiologically relevant dissociation of peptide from the complete class I MHC complex.
Table III summarizes complete data for measurements at 37°C, as well as measurements for peptides with fluorescein-conjugated lysine substitutions at Gln4 and Phe7. From Table III, it is clear that the position of the fluorescein label can influence the measured off-rates, with substitutions at Phe7 differing substantially from those at Val5 and Gln4. However, the effect of the T2M substitution is independent of the position of the fluorescein label, as the rate of peptide dissociation from the fully assembled complex at physiological temperature (k1 in Table III) is a relatively constant 6- to 8-fold slower when methionine is present at position 2 compared with threonine.
Table III.
Kinetic data for the dissociation of fluorescein-labeled peptides from HLA-A2 at 37°C
| Phase 1—Peptide Dissociation from Heterotrimer |
Phase 2—Peptide Dissociation from H Chain/Peptide Heterodimer |
|||||
|---|---|---|---|---|---|---|
| Percent amplitude |
k1 (s−1) | t1/2 (hours) | Fold Rate Decrease for T2M Substitution |
Percent amplitude |
k2 (s−1) | |
| Fluorescein at position 5 | ||||||
| Native | 90 ± 1 | 5.1 ± 0.2 × 10−5 | 3.8 ± 0.2 | 10 ± 2 | 6.2 ± 2.1 × 10−4 | |
| T2M-mod | 87 ± 3 | 7.9 ± 1.0 × 10−6 | 24.5 ± 3.1 | 6.4 ± 0.9 | 13 ± 2 | 1.5 ± 0.8 × 10−4 |
| Fluorescein at position 4 | ||||||
| Native | 89 ± 1 | 7.0 ± 0.2 × 10−5 | 2.7 ± 0.1 | 11 ± 6 | 1.3 ± 0.9 × 10−3 | |
| T2M-mod | 69 ± 7 | 8.7 ± 1.2 × 10−6 | 22.1 ± 3.0 | 8.0 ± 1.2 | 31 ± 7 | 4.5 ± 0.9 × 10−5 |
| Fluorescein at position 7 | ||||||
| Native | 82 ± 2 | 1.7 ± 0.1 × 10−4 | 1.2 ± 0.1 | 18 ± 2 | 8.3 ± 1.9 × 10−4 | |
| T2M-mod | 100 | 2.2 ± 0.1 × 10−5 | 8.8 ± 0.4 | 7.3 ± 0.7 | ND | |
Fig. 7 shows Eyring plots for the data in Fig. 6. Via transition state theory, analysis of kinetic data in this fashion permits determination of activation thermodynamics, with the slope of the line proportional to the activation enthalpy (ΔH‡) and the intercept proportional to the activation entropy (ΔS‡) (see Ref. 23). The thermodynamic parameters derived from this analysis are shown in Table IV. The most important parameters are the differences between the native and T2M-modified peptides (ΔΔH‡ and ΔΔS‡). The values indicate that the slower dissociation of the T2M-modified peptide results from a more unfavorable activation entropy change (ΔΔS‡ < 0), but this is offset by a favorable activation enthalpy change (ΔΔH‡ < 0). As discussed further below, these data are most consistent with the increased hydrophobicity of the T2M-modified peptide (resulting in the unfavorable ΔΔS‡) and the loss of a stabilizing hydrogen bond between Thr2 and Glu63 (resulting in the favorable ΔΔH‡).
FIGURE 7.
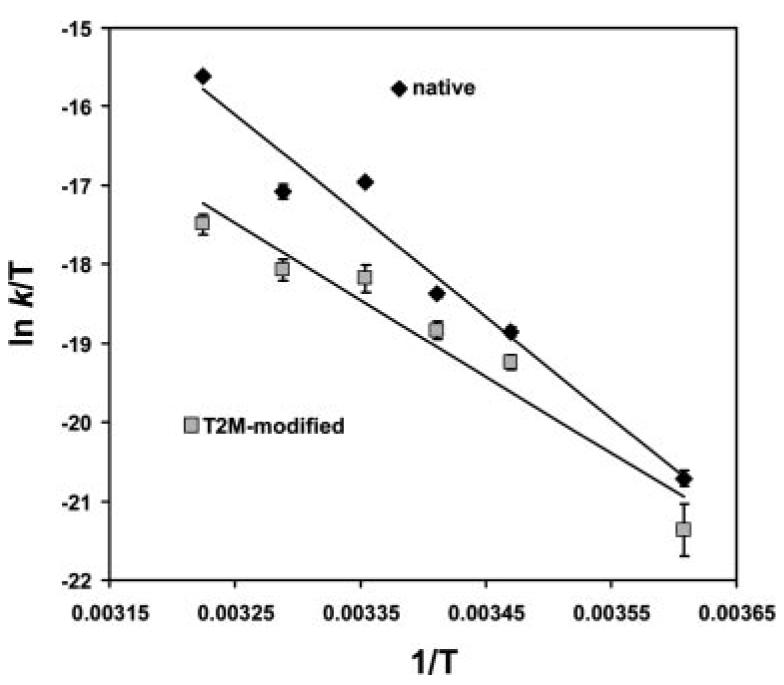
Eyring plots for the peptide dissociation data indicate the thermodynamic basis for the enhanced stability of the gp100209–2M/HLA-A2 complex. Slopes of these lines are proportional to the enthalpic barriers for dissociation, whereas intercepts are proportional to entropic barriers. The steeper slope for the native peptide indicates a more unfavorable enthalpic barrier for dissociation, whereas the greater intercept indicates a more favorable entropic barrier. As the entropic component is larger, the native peptide dissociates faster.
Table IV.
Activation energies for dissociation of the native gp100209 peptide and the T2M-modified variant from HLA-A2a
| ΔH‡ | ΔS‡ | ΔΔH‡ | ΔΔS‡ | |
|---|---|---|---|---|
| Native | 25.4 ± 2.4 | 3.3 ± 0.5 | −6.2 ± 3.5b | −22.7 ± 5.8b |
| T2M-mod | 19.3 ± 2.5 | −19.4 ± 5.8 |
Enthalpies in kilocalories per mole; entropies in calories per mole per K.
Values for the native peptide subtracted from the values for the T2M-modified variant
The crystals of the two complexes were grown from drops at pH 6.5, yet the measurements above were performed at pH 7.4. To control for the effect of pH, we repeated the measurements with the two peptides labeled at position 5 at pH 6.5, 37°C (data not shown). Under these conditions, the half-life of the T2M-modified peptide/MHC complex is ∼5-fold longer than the half-life of the native peptide/MHC complex, compared with ∼7-fold longer at pH 7.4. These results indicate that while pH does influence peptide dissociation rates, the slower dissociation of the T2M-modified gp100209 peptide is unlikely to arise from differences in the two structures at pH 7.4 that are not visible at pH 6.5, where the two complexes were crystallized.
Discussion
The improved immunogenicity of gp100209–2M correlates with the enhanced stability of the peptide/HLA-A2 complex
The crystallographic structures of the native and T2M-modified gp100209 peptides bound to HLA-A2 indicate that the T2M substitution in gp100209 does not alter the structure of the peptide/HLA-A2 complex. In accordance with the structures, in PBMC from 95 individuals, we were unable to determine different frequencies of T cells reactive toward either the native or T2M-modified peptides. Our findings strongly support the interpretation that the enhanced immunogenicity of the T2M-modified peptide is attributable to its enhanced affinity for HLA-A2, rather than TCR cross-reactivity or qualitatively different T cell responses (e.g., partial agonism or antagonism) resulting from different peptide conformations (note that as the T2M substitution in gp100209 does not change the surface that is presented to TCRs, our data imply that Ag-specific TCRs would be unable to distinguish between the native and modified gp100209/HLA-A2 complexes, i.e., TCR binding affinity and kinetics for the gp100209/HLA-A2 complex should be no different from the values for the gp100209–2M/HLA-A2 complex).
As the ∼9-fold enhancement in peptide binding affinity is composed of a ∼7-fold decrease in peptide dissociation rate, our data are most consistent with the increase in immunogenicity resulting from the slower dissociation rate of the T2M-modified peptide. Although the predicted small increase in peptide association rate could also play a role in influencing immunogenicity, the ∼7-fold greater half-life of the complex with the native peptide indicates that in the time for 50% of the modified peptide to dissociate from HLA-A2, only ∼0.8% of the native peptide would remain bound. The greater numbers of gp100209–2M/HLA-A2 complexes on the surfaces of APCs could thus yield a more stable immunological synapse with Ag-specific T cells, leading to more productive signaling.
Comparison with other studies of the effects of the T2M-substitution in gp100209
Our results contradict those of Clay et al. and Denkberg et al. (44, 45), who concluded that significant structural changes result from the T2M substitution in the gp100209 peptide. Clay et al. identified T cell “cloids” that were capable of recognizing the T2M-modified but not the native gp100209 peptide presented by HLA-A2. In light of our data, the findings of Clay et al. may be best attributable to TCR affinity or avidity: T cells expressing TCRs with weak affinity for the native gp100209 peptide presented by HLA-A2 may not respond well to the native peptide as the fast peptide dissociation rate results in too few TCR-peptide/MHC interactions to support the formation of a stable immunological synapse. Conversely, with the T2M-modified peptide, the increased density of peptide/MHC complexes on the APC cell surface increases the likelihood that sufficient TCR-peptide/MHC interactions will occur, despite weak TCR affinity. This interpretation is supported by the experiments of Yu et al. (10), where CTL recognition of APC pulsed with the native gp100209 peptide was substantially reduced when a significant time delay was introduced into the experiment, whereas the those pulsed with the T2M-modified peptide were unaffected.
Denkberg et al. based their conclusions of structural alterations resulting from the T2M modification on an Ab (1A9) that could recognize the T2M-modified but not the native gp100209 peptide bound to HLA-A2 (45). These experiments, which were unlikely to be influenced by the more rapid dissociation of the native peptide, are clearly at odds with our findings. As indicated above, there are no physical differences in the surfaces of the gp100209 and gp100209–2M/HLA-A2 molecules, and electrostatic calculations of surface potentials do not show any effects stemming from removal of the hydrogen bond made from Thr2 to Glu63 (calculations not shown). The temperature factors in the two structures do not suggest different levels of motion between the two peptides. It is conceptually possible that there are differences not visible in the two structures even at the relatively high resolution of 1.8 Å, but if so, this would imply a level of discrimination not previously observed in Ab-Ag recognition. Although it is thus impossible to tell from the structures what, if any, differences the 1A9 Ab may be discerning, we do note that other studies have shown clear differences between Ab and TCR recognition of peptide/MHC complexes (46). Finally, we note the possibility that, as with any x-ray structure, the ensemble-averaged conformation present in vivo may differ from what is observed crystallographically. However, from the data presented in this study, there is no reason to expect that any conformational differences between in vivo and in vitro structures should differ between the complexes of gp100209 and gp100209–2M with HLA-A2.
Implications for vaccine design
The structure with the native gp100209 peptide indicates how the polar threonine side chain is accommodated in the HLA-A2 P2 pocket, forming a hydrogen bond with Glu63 of the HLA-A2 α1 helix. Given the polar environment around the rim of the pocket (Fig. 4), the hydrogen bond made to Glu63 is likely to be strongly stabilizing, as there should be little desolvation penalty for formation of this hydrogen bond. This interpretation is supported by the thermodynamic measurements summarized in Table IV: the native peptide has a much higher enthalpic barrier for dissociation than the T2M-modified peptide, consistent with the need to break a strong hydrogen bond between Thr2 and Glu63, which is not present in the structure with the T2M-modified peptide. Dissociation of the T2M-modified peptide is unfavorable entropically, consistent with the greater hydrophobicity of methionine over threonine (47). Thus, hydrophobicity is the likely reason for the increased stability (and thus immunogenicity) of the gp100209–2M/HLA-A2 complex (although side chain conformational entropy could play a role, its contribution is expected to be small relative to hydrophobicity; Refs. 48 and 49). However, the increase in hydrophobicity is offset considerably by the loss of the Thr2-Glu63 hydrogen bond, accounting for the relatively small ∼9-fold enhancement in binding affinity for the T2M-modified peptide (5) (one caveat to this thermodynamic argument is we have measured activation energies for dissociation, rather than complete binding thermodynamics).
When the native and T2M-modified structures are superimposed, there is no overlap of the β-hydroxyl group of threonine at position 2 of the native peptide with any of the atoms of the methionine in the modified peptide (Fig. 2C). This suggests that it should be possible to achieve further gains in peptide/MHC stability through addition of a β-hydroxyl moiety to methionine—essentially crossing threonine and methionine, producing a β-hydroxy-methionine amino acid capable of hydrogen bonding to Glu63. As there are no structural differences induced by replacement of threonine with methionine, it is reasonable to hypothesize that the structure of gp100209 with β-hydroxy-methionine at position 2 should likewise be unchanged. Thus, our results suggest an approach for engineering the gp100209 peptide to achieve further gains in peptide/MHC stability and, potentially, immunogenicity. Such an approach may also be applicable to other TAA presented by HLA-A2, particularly those for which anchor-fixing using standard amino acids has failed to achieve immunologically significant gains.
In conclusion, we have shown that the T2M substitution in the gp100209 TAA does not alter the structure of the gp100209/HLA-A2 complex. Consistent with this finding, we were unable to identify higher frequencies of T cells specific for either the native or modified peptide in PBMC from a large number of human subjects. These data strongly support the conclusion that the greater immunogenicity of the gp100209–2M peptide is due to the enhanced stability of the peptide/MHC complex, validating the anchor-fixing approach for generating therapeutic vaccine candidates. Finally, the structural and thermodynamic data suggest a route for generating improved APL vaccine candidates using nonstandard amino acids.
Acknowledgments
We thank Drs. Jue Chen and Eva Skrzypczak-Jankun, John Clemens, Tommy Schad, the staff of the Argonne Structural Biology Center, and Joe Miller of Shamrock Structures for assistance.
Footnotes
This work was supported in part by the University of Notre Dame and Grant GM067079 (to B.M.B.) from the National Institute of General Medical Sciences, National Institutes of Health.
Abbreviations used in this paper: TAA, tumor-associated Ag; APL, altered peptide ligand, P2, peptide position 2; RMSD, root mean square deviation; β2m, β2-microglobulin.
Disclosures
B. M. Baker has a patent pending for the use of β-hydroxy amino acids at the first primary anchor for increasing peptide/MHC-I stability.
References
- 1.Novellino L, Castelli C, Parmiani G. A listing of human tumor antigens recognized by T cells: March 2004 update. Cancer Immunol. Immunother. 2005;54:187. doi: 10.1007/s00262-004-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenberg SA, Yang JC, Schwartzentruber DJ, Hwu P, Marincola FM, Topalian SL, Restifo NP, Dudley ME, Schwarz SL, Spiess PJ, et al. Immunologic and therapeutic evaluation of a synthetic peptide vaccine for the treatment of patients with metastatic melanoma. Nat. Med. 1998;4:321. doi: 10.1038/nm0398-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dyall R, Bowne WB, Weber LW, LeMaoult J, Szabo P, Moroi Y, Piskun G, Lewis JJ, Houghton AN, Nikolic-Zugic J. Heteroclitic immunization induces tumor immunity. J. Exp. Med. 1998;188:1553. doi: 10.1084/jem.188.9.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slansky JE, Rattis FM, Boyd LF, Fahmy T, Jaffee EM, Schneck JP, Margulies DH, Pardoll DM. Enhanced antigen-specific antitumor immunity with altered peptide ligands that stabilize the MHC-peptide-TCR complex. Immunity. 2000;13:529. doi: 10.1016/s1074-7613(00)00052-2. [DOI] [PubMed] [Google Scholar]
- 5.Parkhurst M, Salgaller M, Southwood S, Robbins P, Sette A, Rosenberg S, Kawakami Y. Improved induction of melanoma-reactive CTL with peptides from the melanoma antigen gp100 modified at HLA-A*0201-binding residues. J. Immunol. 1996;157:2539. [PubMed] [Google Scholar]
- 6.Kersh GJ, Miley MJ, Nelson CA, Grakoui A, Horvath S, Donermeyer DL, Kappler J, Allen PM, Fremont DH. Structural and functional consequences of altering a peptide MHC anchor residue. J. Immunol. 2001;166:3345. doi: 10.4049/jimmunol.166.5.3345. [DOI] [PubMed] [Google Scholar]
- 7.Sharma AK, Kuhns JJ, Yan S, Friedline RH, Long B, Tisch R, Collins EJ. Class I major histocompatibility complex anchor substitutions alter the conformation of T cell receptor contacts. J. Biol. Chem. 2001;276:21443. doi: 10.1074/jbc.M010791200. [DOI] [PubMed] [Google Scholar]
- 8.Baker BM, Gagnon SJ, Biddison WE, Wiley DC. Conversion of a T cell antagonist into an agonist by repairing a defect in the TCR/peptide/MHC interface: implications for TCR signaling. Immunity. 2000;13:475. doi: 10.1016/s1074-7613(00)00047-9. [DOI] [PubMed] [Google Scholar]
- 9.Hemmer B, Stefanova I, Vergelli M, Germain RN, Martin R. Relationships among TCR ligand potency, thresholds for effector function elicitation, and the quality of early signaling events in human T cells. J. Immunol. 1998;160:5807. [PubMed] [Google Scholar]
- 10.Yu Z, Theoret MR, Touloukian CE, Surman DR, Garman SC, Feigenbaum L, Baxter TK, Baker BM, Restifo NP. Poor immunogenicity of a self/tumor antigen derives from peptide/MHC-I instability and is independent of tolerance. J. Clin. Invest. 2004;114:551. doi: 10.1172/JCI21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irvine KR, Parkhurst MR, Shulman EP, Tupesis JP, Custer M, Touloukian CE, Robbins PF, Yafal AG, Greenhalgh P, Sutmuller RPM, et al. Recombinant virus vaccination against “self” antigens using anchor-fixed immunogens. Cancer Res. 1999;59:2536. [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg SA, Yang JC, Schwartzentruber DJ, Hwu P, Topalian SL, Sherry RM, Restifo NP, Wunderlich JR, Seipp CA, Rogers-Freezer L, et al. Recombinant fowlpox viruses encoding the anchor-modified gp100 melanoma antigen can generate antitumor immune responses in patients with metastatic melanoma. Clin. Cancer Res. 2003;9:2973. [PMC free article] [PubMed] [Google Scholar]
- 13.Garboczi DN, Hung DT, Wiley DC. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl. Acad. Sci. USA. 1992;89:3429. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 16.Bouvier M, Guo HC, Smith KJ, Wiley DC. Crystal structures of HLA-A*0201 complexed with antigenic peptides with either the amino- or carboxyl-terminal group substituted by a methyl group. Proteins. 1998;33:97. doi: 10.1002/(sici)1097-0134(19981001)33:1<97::aid-prot9>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 17.Murshudov G, Vagin A, Dodson E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. Biol. Crystallogr. 1997;53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 18.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP and automatic interpretation of protein electron density maps. Macromolec. Crystallogr. D. 2003;374:229. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 19.McRee DE. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 20.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283. [Google Scholar]
- 21.Rodriguez R, Chinea G, Lopez N, Pons T, Vriend G. Homology modeling, model and software evaluation: three related resources. Bioinformatics. 1998;14:523. doi: 10.1093/bioinformatics/14.6.523. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski RA. Surfnet—a program for visualizing molecular-surfaces, cavities, and intermolecular interactions. J. Mol. Graphics. 1995;13:323. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 23.Binz AK, Rodriguez RC, Biddison WE, Baker BM. Thermodynamic and kinetic analysis of a peptide-class I MHC interaction highlights the noncovalent nature and conformational dynamics of the class I heterotrimer. Biochemistry. 2003;42:4954. doi: 10.1021/bi034077m. [DOI] [PubMed] [Google Scholar]
- 24.Bevington PR, Robinson DK. Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill; New York: 1992. [Google Scholar]
- 25.Madden DR, Garboczi DN, Wiley DC. The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell. 1993;75:693. doi: 10.1016/0092-8674(93)90490-h. [Published erratum appears in 1994 Cell 76:following 410.] [DOI] [PubMed] [Google Scholar]
- 26.Khan AR, Baker BM, Ghosh P, Biddison WE, Wiley DC. The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J. Immunol. 2000;164:6398. doi: 10.4049/jimmunol.164.12.6398. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Turner R, Baker BM, Biddison WE. MHC allele-specific molecular features determine peptide/HLA-A2 conformations that are recognized by HLA-A2-restricted T cell receptors. J. Immunol. 2002;169:3146. doi: 10.4049/jimmunol.169.6.3146. [DOI] [PubMed] [Google Scholar]
- 28.Gagnon SJ, Wang Z, Turner R, Damirjian M, Biddison WE. MHC recognition by hapten-specific HLA-A2-restricted CD8+ CTL. J. Immunol. 2003;171:2233. doi: 10.4049/jimmunol.171.5.2233. [DOI] [PubMed] [Google Scholar]
- 29.Collins EJ, Garboczi DN, Wiley DC. Three-dimensional structure of a peptide extending from one end of a class I MHC binding site. Nature. 1994;371:626. doi: 10.1038/371626a0. [DOI] [PubMed] [Google Scholar]
- 30.Gao GF, Tormo J, Gerth UC, Wyer JR, McMichael AJ, Stuart DI, Bell JI, Jones EY, Jakobsen BK. Crystal structure of the complex between human CD8α(α) and HLA-A2. Nature. 1997;387:630. doi: 10.1038/42523. [DOI] [PubMed] [Google Scholar]
- 31.Hillig RC, Coulie PG, Stroobant V, Saenger W, Ziegler A, Hulsmeyer M. High-resolution structure of HLA-A*0201 in complex with a tumour-specific antigenic peptide encoded by the MAGE-A4 gene. J. Mol. Biol. 2001;310:1167. doi: 10.1006/jmbi.2001.4816. [DOI] [PubMed] [Google Scholar]
- 32.Kirksey TJ, Pogue-Caley RR, Frelinger JA, Collins EJ. The structural basis for the increased immunogenicity of two HIV-reverse transcriptase peptide variant/class I major histocompatibility complexes. J. Biol. Chem. 1999;274:37259. doi: 10.1074/jbc.274.52.37259. [DOI] [PubMed] [Google Scholar]
- 33.Kuhns JJ, Batalia MA, Yan S, Collins EJ. Poor binding of a HER-2/neu epitope (GP2) to HLA-A2.1 is due to a lack of interactions with the center of the peptide. J. Biol. Chem. 1999;274:36422. doi: 10.1074/jbc.274.51.36422. [DOI] [PubMed] [Google Scholar]
- 34.Sliz P, Michielin O, Cerottini J-C, Luescher I, Romero P, Karplus M, Wiley DC. Crystal structures of two closely related but antigenically distinct HLA-A2/melanocyte-melanoma tumor-antigen peptide complexes. J. Immunol. 2001;167:3276. doi: 10.4049/jimmunol.167.6.3276. [DOI] [PubMed] [Google Scholar]
- 35.Zhao R, Loftus DJ, Appella E, Collins EJ. Structural evidence of T cell xeno-reactivity in the absence of molecular mimicry. J. Exp. Med. 1999;189:359. doi: 10.1084/jem.189.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reid SW, McAdam S, Smith KJ, Klenerman P, O'Callaghan CA, Harlos K, Jakobsen BK, McMichael AJ, Bell JI, Stuart DI, Jones EY. Antagonist HIV-1 gag peptides induce structural changes in HLA B8. J. Exp. Med. 1996;184:2279. doi: 10.1084/jem.184.6.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JK, Stewart-Jones G, Dong T, Harlos K, Di Gleria K, Dorrell L, Douek DC, van der Merwe PA, Jones EY, McMichael AJ. T cell cross-reactivity and conformational changes during TCR engagement. J. Exp. Med. 2004;200:1455. doi: 10.1084/jem.20041251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webb AI, Dunstone MA, Chen W, Aguilar M-I, Chen Q, Jackson H, Chang L, Kjer-Nielsen L, Beddoe T, McCluskey J, et al. Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue. J. Biol. Chem. 2004;279:23438. doi: 10.1074/jbc.M314066200. [DOI] [PubMed] [Google Scholar]
- 39.Ding YH, Baker BM, Garboczi DN, Biddison WE, Wiley DC. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity. 1999;11:45. doi: 10.1016/s1074-7613(00)80080-1. [DOI] [PubMed] [Google Scholar]
- 40.Eriksson AE, Baase WA, Zhang XJ, Heinz DW, Blaber M, Baldwin EP, Matthews BW. Response of a protein-structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255:178. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 41.Baxter TK, Gagnon SJ, Davis-Harrison RL, Beck JC, Binz A-K, Turner RV, Biddison WE, Baker BM. Strategic mutations in the class I MHC HLA-A2 independently affect both peptide binding and T cell receptor recognition. J. Biol. Chem. 2004;279:29175. doi: 10.1074/jbc.M403372200. [DOI] [PubMed] [Google Scholar]
- 42.Gakamsky DM, Boyd LF, Margulies DH, Davis DM, Strominger JL, Pecht I. An allosteric mechanism controls antigen presentation by the H-2Kb complex. Biochemistry. 1999;38:12165. doi: 10.1021/bi9905821. [DOI] [PubMed] [Google Scholar]
- 43.Gakamsky DM, Davis DM, Strominger JL, Pecht I. Assembly and dissociation of human leukocyte antigen (HLA)-A2 studied by real-time fluorescence resonance energy transfer. Biochemistry. 2000;39:11163. doi: 10.1021/bi000763z. [DOI] [PubMed] [Google Scholar]
- 44.Clay TM, Custer MC, McKee MD, Parkhurst M, Robbins PF, Kerstann K, Wunderlich J, Rosenberg SA, Nishimura MI. Changes in the fine specificity of gp100209–217-reactive T cells in patients following vaccination with a peptide modified at an HLA-A2.1 anchor residue. J. Immunol. 1999;162:1749. [PubMed] [Google Scholar]
- 45.Denkberg G, Klechevsky E, Reiter Y. Modification of a tumor-derived peptide at an HLA-A2 anchor residue can alter the conformation of the MHC-peptide complex: probing with TCR-like recombinant antibodies. J. Immunol. 2002;169:4399. doi: 10.4049/jimmunol.169.8.4399. [DOI] [PubMed] [Google Scholar]
- 46.Biddison WE, Turner RV, Gagnon SJ, Lev A, Cohen CJ, Reiter Y. Tax and M1 peptide/HLA-A2-specific Fabs and T cell receptors recognize nonidentical structural features on peptide/HLA-A2 complexes. J. Immunol. 2003;171:3064. doi: 10.4049/jimmunol.171.6.3064. [DOI] [PubMed] [Google Scholar]
- 47.Cornette JL, Cease KB, Margalit H, Spouge JL, Berzofsky JA, DeLisi C. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J. Mol. Biol. 1987;195:659. doi: 10.1016/0022-2836(87)90189-6. [DOI] [PubMed] [Google Scholar]
- 48.Lee KH, Xie D, Freire E, Amzel LM. Entropy changes in biological processes—side-chain entropy losses in folding and binding. Biophys. J. 1994;66:A13. [Google Scholar]
- 49.Baker BM, Murphy KP. Prediction of binding energetics from structure using empirical parameterization. Methods Enzymol. 1998;295:294. doi: 10.1016/s0076-6879(98)95045-5. [DOI] [PubMed] [Google Scholar]


