Abstract
RNA oligomers of length 40–60 nt can self-assemble into covalent versions of the Azoarcus group I intron ribozyme. This process requires a series of recombination reactions in which the internal guide sequence of a nascent catalytic complex makes specific interactions with a complement triplet, CAU, in the oligomers. However, if the CAU were mutated, promiscuous self-assembly may be possible, lessening the dependence on a particular set of oligomer sequences. Here, we assayed whether oligomers containing mutations in the CAU triplet could still self-construct Azoarcus ribozymes. The mutations CAC, CAG, CUU and GAU all inhibited self-assembly to some degree, but did not block it completely in 100 mM MgCl2. Oligomers containing the CAC mutation retained the most self-assembly activity, while those containing GAU retained the least, indicating that mutations more 5′ in this triplet are the most deleterious. Self-assembly systems containing additional mutant locations were progressively less functional. Analyses of properly self-assembled ribozymes revealed that, of two recombination mechanisms possible for self-assembly, termed ‘tF2’ and ‘R2F2’, the simpler one-step ‘tF2’ mechanism is utilized when mutations exist. These data suggest that self-assembling systems are more facile than previously believed, and have relevance to the origin of complex ribozymes during the RNA World.
INTRODUCTION
The emergence of an RNA World required the advent of RNA-like polymers of sufficient length to possess catalytic activities. In general, while short RNA motifs can catalyze simple, entropically favorable reactions such as phosphoester bond self-cleavage, more complex reactions require RNAs of the modern tRNA size (∼75 nt) or greater. These reactions include ligation, phosphotransfer and polymerization, in which more than one substrates is involved and/or multiple sequential chemical steps are needed. Every known non-self-cleaving ribozyme contains several stem-loop structures, and many of the larger catalysts require pseudoknots. Abiotic syntheses of RNA oligomers from activated monomers has been demonstrated, but the upper limit of polymer length seen so far has been 50–60 nt, and yields are biased towards much shorter products (1–3).
One means by which polymer length could increase is recombination, the energy-neutral disproportionation of shorter oligomers. A metathesis reaction between, say, two 10-mers could generate an 18-mer and a 2-mer if it were to proceed in a highly asymmetrical fashion. In principle this can happen via either an uncatalyzed or a catalyzed pathway. The reactions reported by Chetverin and colleagues (4,5) are potential examples of the former. In this case, data can be interpreted to support a mechanism by which two RNA strands partially hybridize through base pairing of short, complementary regions, producing a staggered complex in which the free 3′-OH of one strand attacks a phosphate group on the opposing strand. This would result in recombinant molecules of potentially vastly different lengths than their progenitors. Although these authors could not unambiguously rule out involvement by protein enzymes, recent data by Lutay et al. (6) support the notion that RNA species themselves can be responsible for the reaction.
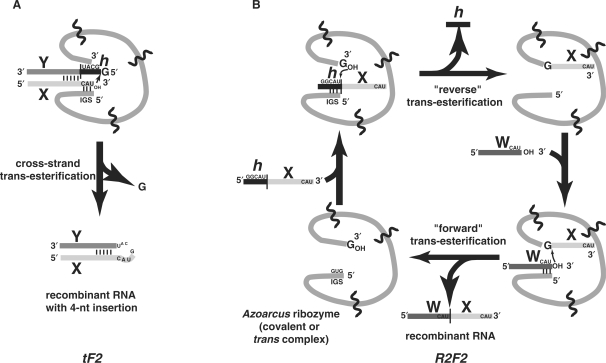
On the other hand, it is quite clear that group I and group II ribozymes can catalyze RNA recombination (7–10). These reactions proceed with the general form A·B + C·D → C·B + A·D. One manner in which this can take place is in a single step involving a cross-strand nucleophilic attack (analogous to the putative non-catalyzed reaction), as seen with the Anabaena ribozyme (11). We will refer to this mechanism as the ‘tF2’ reaction, because it is chemically analogous in the forward direction to the second step of in vivo splicing of tRNA by group I introns: the exon-ligation step to produce the anticodon loop. Alternatively, a more complex mechanism involving two sequential phosphotransfer reactions can be catalyzed by the Azoarcus and Tetrahymena group I introns (10) as well as by group II introns (9). We will refer to this mechanism as the ‘R2F2’ reaction, because it is chemically analogous to the reverse direction followed by the forward direction of in vivo splicing by group I introns, and is general enough to accommodate substrates that are not complementary to each other (10). Both the tF2 and R2F2 reactions are formally recombinations when all substrates and products are considered, and both mimic in vivo reactions occurring at the 3′ splice site (Figure 1).
Figure 1.
The two possible mechanisms of RNA-catalyzed RNA recombination. (A) The ‘tF'2’ mechanism in which the ribozyme, which can either be a covalently contiguous Azoarcus RNA or a trans complex comprised of 2–4 hydrogen-bonded RNA fragments (delineated by wavy lines), binds a duplex RNA and catalyzes a cross-strand attack resulting in a short insertion that is usually GCAU (12). Slippage of the duplex in the active site can lead to shorter (e.g. CAU) or longer (e.g. GGCAU) insertions. This reaction occurs in the forward direction during the second step of tRNA self-splicing (hence tF2) and was first observed in the Anabaena group I intron (11). (B) The ‘R2F2’ mechanism in which the ribozyme catalyzes two successive phosphotransfer reactions on exogenous RNA substrates resulting in a recombinant RNA molecule (10). Correct splicing events are guided by a specific 3 bp interaction between the IGS (GUG) in the catalyst and its complement (CAU) in the substrates. This reaction is effectively the reverse of the second step of in vivo group I intron splicing followed by the forward of the second step (hence R2F2).
We have shown how RNA recombination can result in the spontaneous construction of covalently contiguous group I ribozymes from oligomers of length 40–60 nt in the absence of any pre-existing ribozyme (12). Four fragments, W, X, Y and Z of the ∼200 nt Azoarcus ribozyme will self-assemble into a catalytically active trans complex, which can then perform a series of recombination reactions using additional copies of the fragments as substrates, ultimately producing covalent versions of the ribozyme (Figure 2). These products appear to catalyze further ribozyme construction, suggesting an autocatalytic component to the system (12). This provides a good model for the early stages of the RNA World in which genetic information needs to bootstrap itself up from chaos (13,14). In particular, the system is ostensibly dependent on the presence of an internal guide sequence (IGS) in the 5′ portion of the catalyst, as this interacts with its complement to guide the correct series of recombinations among fragments (Figures 1 and 2). In the Azoarcus RNA, the IGS is GUG, and its complement is 5′–CAU–3′, because group I introns are characterized as requiring a G–U wobble interaction at the splice site. Consequently the CAU triplets serve as ‘tag’ sequences, directing catalysts where to splice, under either the tF2 or the R2F2 mechanisms.
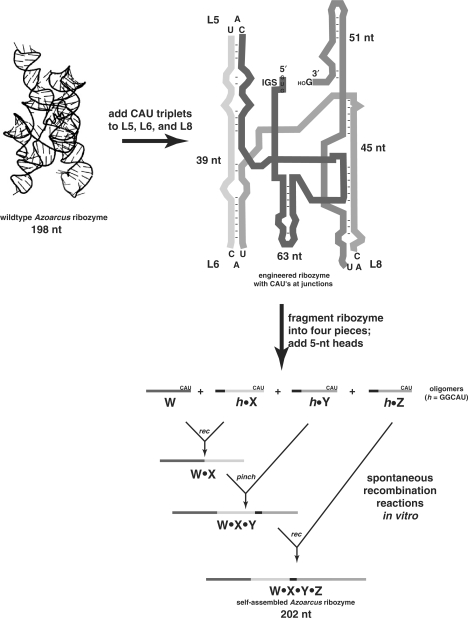
Figure 2.
Design of the Azoarcus RNA self-assembly system. The wild-type L8 Azoarcus ribozyme (197 nt) was modified by the insertion of CAU triplets in three loop regions. Insertions in L5 and L8 were created by two single-nucleotide substitutions each, while that in L6 required a single-nucleotide insertion as well, resulting in a 198 nt construct (12). Four fragments of the ribozyme were made by partitioning this construct immediately 3′ of the CAU triplets and adding 5 nt head regions (GGCAU) on the 5′ ends of three of them, creating oligomers W (63 nt), h·X (5 + 39 = 44 nt), h·Y (5 + 45 = 50 nt), and h·Z (5 + 51 = 56 nt). When these four oligomers are mixed together in an equimolar ratio at 48°C in 100 mM MgCl2, they can spontaneously self-assemble into a covalently contiguous ∼200 nt ribozyme by a series of recombination reactions (12).
Here, we sought to investigate the details of the self-assembly pathways. (Throughout this article, we will refer to the autonomous self-construction of covalently contiguous RNAs as ‘self-assembly’, although we acknowledge that this term is often used to refer to non-covalent macromolecular aggregation events, including the formation of ribozymes in trans.) Specifically, we were interested in testing the hypothesis that self-assembly would still be feasible in the Azoarcus system if the tag sequences were mutated away from CAU. This would portend a broader generality of this self-assembly mechanism beyond that of a specific set of RNA oligomers. Our results show that in 100 mM MgCl2, many mutations of this sequence do in fact allow the system to retain its self-assembly property. Auspiciously, the specific effects of the triplet sequence identity and location(s) in the original oligomers reveal a great deal about the range of mechanisms available to ribozyme self-construction.
MATERIALS AND METHODS
RNAs were either purchased from IDT (oligomer NNN and all-CAU-containing oligomers h·X, h·Y and h·Z) or prepared by run-off transcription from double-stranded DNA templates constructed through recursive gene synthesis (all other oligomers). All RNAs were gel purified and desalted prior to use. Salts and buffers were made from the highest purity available (Sigma-Aldrich) and all water used was nuclease free (Ambion). Barrier pipette tips and other strict contamination controls were employed at all times.
Self-assembly experiments
Ribozyme covalent self-assembly from four oligomers was performed as described previously (12). Briefly, RNA oligomers were incubated together at 48°C at a final concentration of 1 μM each. All reactions contained a final concentration of 100 mM MgCl2 and 30 mM Tris (pH 7.5) unless otherwise indicated. Reactions were carried out in triplicate for 6 h in 200 μl or 600 μl microcentrifuge tubes and then quenched by precipitation in ethanol. The RNAs were rehydrated in a gel-loading solution containing 8 M urea, SDS and bromphenol blue dye, heat denatured at 80°C for 4 min, and then immediately electrophoresed through 8% polyacrylamide/8 M urea gels. In most cases, the W fragment was 5′-end-labeled with γ[32P].ATP and OptiKinase (USB) prior to use. This allowed visualization of the products via phosphorimaging with a Typhoon 9200 instrument (GE Healthcare). In some cases no radioactivity was employed, and all oligomers were visualized by SYBR Green II staining, although when quantification was needed, this technique was not used. To differentiate between the two possible recombination mechanisms at the W–X junction, two binary (two-piece) self-assembly reactions were performed by incubating 2 μM W with 2 μM h·X·Y·Z for 3 h at 48°C in 100 mM MgCl2. In parallel reactions, the sequences at the W–X junctions were altered slightly: in one case they matched the four-piece reactions performed elsewhere, and in the other case the CAU sequence was transposed one position to match the construct used at the X–Y junction in which the tF2 mechanism is heavily favored over the R2F2 mechanism.
Genotyping
Full-length RNA covalent constructs were identified by comparison to a bona fide Azoarcus RNA run as a size control. These bands were carefully excised from the gel and subjected to reverse transcription using the primer T20a (5′– CCGGTTTGTGTGACTTTCGCC–3′), which targets the 3′ portion of the Z fragment. One-twentieth of these reactions were used to seed PCR reactions employing T20a and T2.1a (5′–CTGCAGAATTCTAATACGACTCACTATAGTGCCTTGCGCCGGGAA–3′) primers (15), the latter being specific for the 5′ portion of W. The PCR products were cloned into the vector pJet (Fermentas) and transformed into Escherichia coli. Individual colonies were picked as templates for colony PCR reactions employing the primers pJet-F and pJet-R (Fermentas), which generate products of ∼300 bp [= the insert size (∼220 bp) plus ∼80 bp]. Products of the correct size were genotyped using BigDye (v.3) cycle sequencing chemistry and a Prism 3100 (ABI) instrument.
Mis-cleavage assays
Target RNA oligomers for cleavage assays by the Azoarcus ribozyme, SL-1 (5′– GGCAUCUUCGGAUGCAGGGGAGGCAGCUCCCGAUGGAAGUGACG AGCCAGCGUUCUCAACAGUAUUGACUGAACCUAAAAGCCAAUCGCAGGCUCAGC–3′) and NNN (5′–GAAUCUUCGUCAGGAUCCAGCACGUACAACCUAUCAUCG–3′), were designed to include multiple sites where the ribozymes could potential bind via the IGS and perform an endoribonuclease reaction and liberate a 5′ fragment. These substrates were 5′end-labeled using γ[32P].ATP and OptiKinase and gel purified. Cleavage assays were performed by incubating either full-length Azoarcus ribozyme or its four component fragments (W, h·X, h·Y and h·Z) with one of the two substrates under varying MgCl2 concentrations ranging from 4 to 100 mM. In all cases, the final catalyst concentration was 2 μM, the substrate concentration was 0.5 μM, and reactions were carried out in 30 mM Tris (pH 7.5) for 20 min at 48°C. Reactions were quenched as above and electrophoresed on 15% polyacrylamide/8 M urea gels and visualized by phosphorimaging. T1 nuclease and OH− ladders were generated by standard methods and run alongside reaction products.
RESULTS
Canonical (CAU) self-assembly
We first examined the products formed by the canonical four-piece self-assembly reaction reported previously (12) employing the IGS complement of the wild-type Azoarcus ribozyme, CAU. We placed this triplet in the head (h) sequences (GGCAU) that precede the splice site (·) in three of the fragments, h·X, h·Y and h·Z, and at the 3′ ends of all four fragments, W, h·X, h·Y and h·Z. Using 1 µM concentrations of each oligomer incubated at 48°C for 6 h in 100 mM MgCl2 (pH 7.5), we observed 4.9% ± 0.6% (n = 4 trials) self-assembly based on moles of 5′-[32P]-radiolabeled W converted to W·X·Y·Z. We then excised the band from a denaturing polyacrylamide gel corresponding to the full-length W·X·Y·Z product (∼200 nt), amplified via RT-PCR, cloned and obtained nucleotide sequences from 21 individual colonies in order to examine the successful genotypes, paying particular attention to the junction regions formed by recombination between fragments. These sequences are described in Table 1. With the caveat that our RT-PCR primers target molecules bounded by W (5′) and Z (3′), the overwhelming tendency of the system is to generate full-length products containing the W, X, Y and Z fragments assembled in the correct order, as noted before (12). Of the 21 colonies genotyped, 20 were W·X·Y·Z, while only one was not, being W·X followed by internal rearrangements and then a 3′ fragment of Z. No molecules contained sequences such as W·Y·X·Z, which would in theory be possible and possess the same length as Azoarcus RNA.
Table 1.
Inserted sequences at junctions of all-CAU four-piece self-assembly reactions
| Clone number | W–X junction | X–Y junction | Y–Z junction | Notes |
|---|---|---|---|---|
| 688 | GCAU | GCAU | GCAU | tF2 rxn at all three junctions |
| 689 | No insertion | GCAU | GCAUG | |
| 690 | CAU | GCAU | GCAU | |
| 691 | No insertion | GCAU | No insertion | |
| 692 | GCAU | GCAU | No insertion | |
| 693 | GCAU | CAU | GGCAU | |
| 694 | No insertion | GCAU | G | |
| 695 | GCAU | (No insertion) | (No insertion) | WX(Z): incorrect self-assembly |
| 720 | No insertion | GCAU | No insertion | |
| 721 | No insertion | GCAU | GCAU | Also: 1 nt deletion in Z |
| 722 | No insertion | GCAU | GCAU | |
| 724 | No insertion | GCAU | No insertion | |
| 725 | No insertion | GGCAU | No insertion | |
| 726 | No insertion | GCAU | No insertion | |
| 727 | No insertion | GCAU | No insertion | |
| 728 | No insertion | CAU | GCAUG | |
| 729 | No insertion | CAU | No insertion | |
| 730 | No insertion | GCAU | No insertion | |
| 731 | U | CAU | No insertion | |
| 734 | No insertion | GCAU | No insertion | Also: 24 nt deletion in Z |
| 735 | No insertion | CAU | G |
The sequences at the fragment junctions are indicative of which mechanism, tF2 or R2F2, occurred to join the fragments. The original system was designed with the intent that self-assembly would occur entirely through the R2F2 mechanism (12). This would result when the h heads are cleaved off following the CAU in h by catalytic molecules—either trans assemblies or covalent molecules—and then a second fragment is recombined to the first fragment by a subsequent (reverse) transesterification reaction (Figure 1B). This was detected at the W–X and Y–Z junctions by bulk genotyping of the products (12). However, at the X–Y junction, insertions of 3–4 nt are common, and this led us to propose that the tF2 mechanism, requiring only one catalytic event, was occurring at that junction (12). The junction sequences of ‘individual’ molecules in Table 1 confirm these assessments, but also showed that the tF2 reaction can occasionally occur at the W–X and Y–Z junctions as well. At the X–Y junctions, all 20 of the W·X·Y·Z clones possessed a short insertion, which is typically GCAU, but was simply CAU in four cases, and was expanded to GGCAU in one case. At the W–X and Y–Z junctions, insertions indicative of the tF2 reaction occurred 25% and 40% of the time, respectively, while no insertions were seen the remainder of instances, indicative of the R2F2 reaction. Summing over all three junctions, the tF2 frequency was 55% and the R2F2 frequency was 45%, demonstrating that both types of recombination were contributing significantly to autonomous self-assembly of RNA catalysts. Other types of mutations, such as single-nucleotide substitutions, insertions and deletions, were sporadically seen elsewhere in the molecule, but these were very rare.
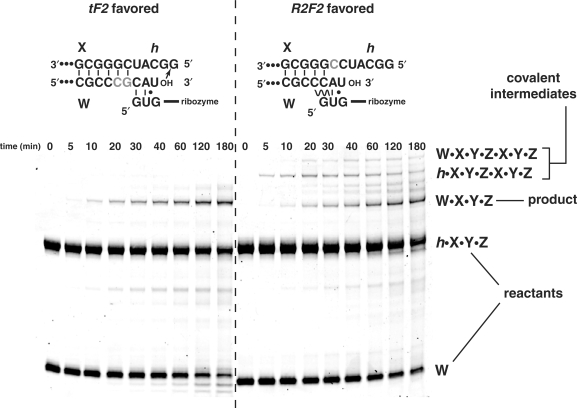
Using two-piece self-assembly reactions, W + h·X·Y·Z, we were able to differentiate between the tF2 and R2F2 mechanisms (Figure 3). The tF2 mechanism should generate ligated products directly, while the R2F2 mechanism should produce covalent intermediates (h·X·Y·Z·X·Y·Z in this case) prior to the appearance of the full-length ribozyme (Figure 1). With the simpler binary reaction we could test for the appearance of such intermediates, and test the effect of moving the CAU at the W–X junction one position 3′, as it exists in the X–Y junction. This shift greatly favors the tF2 mechanism because the C of the CAU in W is free to pair with the IGS instead of with X. With the W–X junction unperturbed, where we detect the R2F2 mechanism 75% of the time in the four-piece reaction (see above), we did in fact observe the 275 nt h·X·Y·Z·X·Y·Z covalent intermediate after only 5 min, while the ribozyme product W·X·Y·Z did not appear until 20 min (Figure 3). In contrast, when the junction was altered to favor the tF2 mechanism, we observed almost no covalent intermediates.
Figure 3.
Confirmation of a covalent intermediate during the R2F2 recombination reaction. The binary reaction W + h·X·Y·Z was carried out using two different constructs at the W–X junction, as shown, and run on a denaturing 8% polyacrylamide gel stained with SYBR Green dye so all RNAs are visible. The construction on the left, created by moving the relative position of the CAU triplets by the addition of the gray nucleotides, mimics that at the X–Y junction in which the tF2 mechanism is highly favored. The construction on the right, used elsewhere in this study, favors the R2F2 mechanism (see text), which produces the covalent 275 nt intermediate h·X·Y·Z·X·Y·Z prior to the ribozyme product W·X·Y·Z, plus the 333 nt covalent intermediate W·X·Y·Z·X·Y·Z after products have been formed.
Promiscuous self-assembly
We then investigated the effects of mutations in the CAU triplets on self-assembly. Our hypothesis was that any mutations away from CAU would disrupt the IGS–IGS complement interaction during catalysis and therefore hinder, but not inhibit the tF2 and the R2F2 mechanisms, both of which should require two Watson–Crick base pairs preceding a terminal G–U wobble pair at the splice site, based on known group I intron function. Promiscuity was plausible because self-assembly reactions are most operative at 48°C in 100 mM MgCl2, permissive conditions that are significantly cooler and more salty than the optimal conditions for in vitro splicing by the Azoarcus ribozyme: 60°C and 15 mM MgCl2 (16). Thus we tested the effects of four different mutations of the CAU triplets on self-assembly: CAC, CAG, CUU and GAU. These mutations can be engineered into between one and six positions of the four-piece self-assembly system: at W* (at the 3′ end of W), *X (in the h segment of X), X* (at the 3′ end of X), *Y (in the h segment of Y), Y* (at the 3′ end of Y) and/or *Z (in the h segment of Z).
The effect of the identity of the mutant sequence
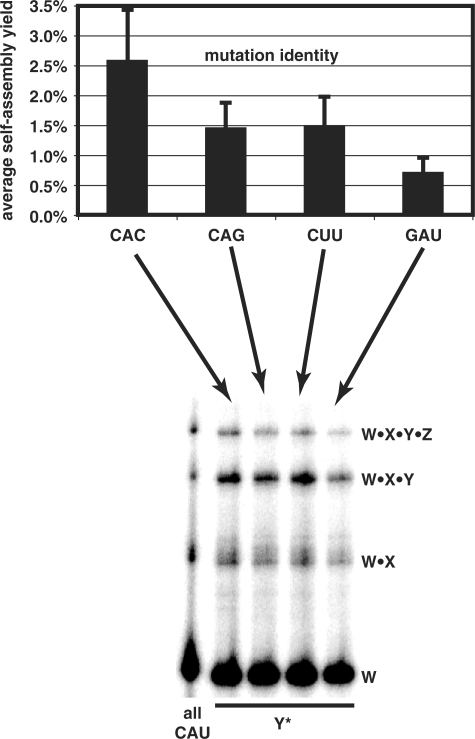
Using the set of standardized self-assembly conditions described above, we observed that mutations away from CAU were in fact tolerated by the system (Figure 4). When a mutation was placed in only one of the six possible locations, all four mutants allowed retention of self-assembly by the four fragments, though the overall yield dropped below that of the all-CAU system. This result verified that promiscuous self-assembly is possible, and variants of the wild-type Azoarcus RNA system can self-assemble, even if the IGS–IGS complement is not a canonical match.
Figure 4.
Effect of various IGS-complement mutations on self-assembly yields. Graph shows average effect (and standard errors) of CAU mutations at a single site after 6 h incubations (n = 18 each: averaged over three independent trails at all six possible sites, W*, *X, X*, *Y, Y* and *Z). Gel shows example run for mutations at Y*.
It is important to note that the yields for the all-CAU self-assembly cannot be strictly compared to those of the mutant self-assemblies. This is a consequence of the fact that the CAU-containing oligomers (with the exception of W) were created by solid-phase synthesis and thus possessed unique 3′ ends, while all mutant oligomers were generated by run-off transcription from PCR products and may have non-uniform 3′ ends. Self-assembly is reliant on a precise 3′-OH nucleophile on the 3′ nt, meaning that the mutant self-assembly reactions are pre-biased to be less productive than the all-CAU reaction. Thus the yields of the mutant reactions are actually conservative. These yield are, however, directly comparable with each other.
In any case, the identity of the mutation clearly had an impact. The triplet CAC was the most tolerated, followed by CAG and CUU, which had roughly equal detrimental effects on self-assembly, and then GAU, which was the least tolerated. The difference between CAC and GAU was statistically significant (t-test, P = 0.05). Thus to a first approximation, mutations in the 3′ nt of the IGS complement are the most tolerated, followed by the middle position and then the 5′ position. We did not, however, test all permutations of CAU to test this trend exhaustively.
The effect of the number of mutant sites
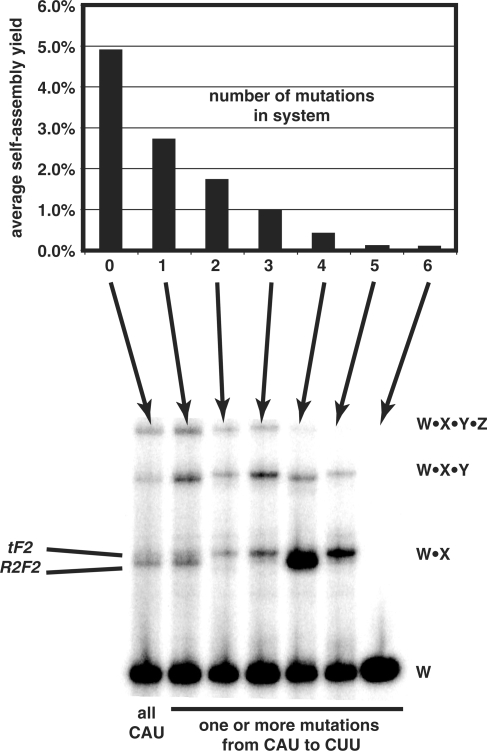
Because each mutation did have a detrimental effect on self-assembly yields, we reasoned that additional mutations would have a cumulative detrimental effect. Thus we tested the impact of engineering in one, two, three, four, five or all six sites in a system with a particular non-CAU triplet. As expected, the more sites mutated, the poorer the self-assembly yield (Figure 5, graph). Using a mutation of moderate effect (CUU) as an example, there is a monotonically decreasing yield with an increase in the number of sites mutated (Figure 5, gel). When all six CAU sites are mutated to CUU, no self-assembly is seen under the standardized assay conditions. Yet with three and even four or five mismatched IGS–IGS complements (GUG with CUU) required along the self-assembly pathway, a detectable yield of full-length product can be observed.
Figure 5.
Effects of multiple IGS-complement mutations on self-assembly yields. Graph shows average effect of self-assembly trials when 0–6 of the CAU sequences are mutated, averaged over all four mutations tested, CAC, CAG, CUU and GAU. Gel shows example of self-assembly reactions using the CUU mutation: lane 1, all CAU (no mutations); lane 2, CUU at *Y; lane 3, CUUs at *X and *Y; lane 4, CUUs at *X, *Y and *Z; lane 5, CUUs at X*, *Y, Y* and *Z, lane 6, CUUs at *X, X*, *Y, Y* and *Z; lane 7, CUUs at W*, *X, X*, *Y, Y* and *Z. Mutations at *X favor the tF2 mechanism at the W–X junction, resulting in an upshift of the W·X intermediate by ∼4 nt.
The effect of the location(s) of the mutant sites
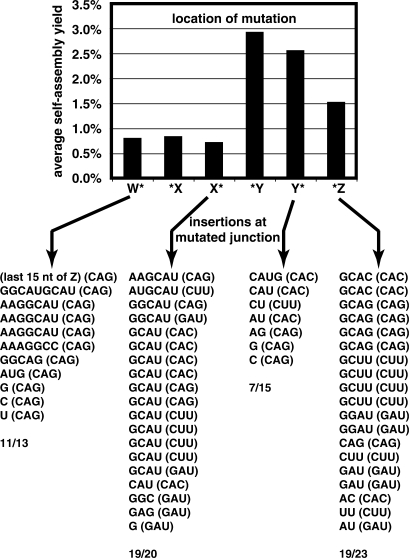
We next tested whether the effect of location (i.e. where, and in which original oligomer) would influence self-assembly yields. To do this, we focused on self-assembly reactions containing only one mutant site, in part because these yields were the highest, and in part because testing all permutations of multiple-site mutations would be prohibitive. For self-assembly reaction yields, we tested all four possible mutations used above (CAC, CAG, CUU and GAU), at all six possible sites, for 24 comparisons altogether. We performed each reaction three times and computed means and standard errors for comparison purposes.
The location of a mutation did have some effect on self-assembly yields (Figure 6). In general, mutations in Y, either *Y or Y*, were well tolerated. Assemblies with CAC and CUU mutations at either location were particularly robust, with yields of 3–7% under the standard assay conditions. Mutations at *Z were the next best tolerated, and in the case of CAG gave yields actually higher than mutations in Y. Finally, mutations in W or at either location in X were the least tolerated, lowering self-assembly yields to 0–2% in all cases. Across all mutations, the only statistically significantly different pair-wise comparisons are *Y versus *X or X* (model I ANOVA with k = 15 planned comparisons of means, P = 0.05). Mutations in W or X are probably the most deleterious because we have previously inferred that the order of assembly most often proceeds 5′ to 3′, W → W·X → W·X·Y → W·X·Y·Z, and thus inhibition of the first recombination reactions in the series would have a greater overall impact on yield (12).
Figure 6.
Effect of location of mutation on self-assembly yields. Graph shows average self-assembly yields when CAU is mutated at only the indicated location in one of the four fragments. Mutations at a particular junction increase the probability that the tF2 mechanism will be used at that junction; the sequences below the graph delineate the observed insertions at particular junctions when a mutation exists before or after that junction (overall insertion frequencies shown at bottom; the mutation identities are given in parentheses). Sequence analyses of junctions were not performed for *X and *Y and only with the CAG mutation at W*.
Sequence analysis of junctions in promiscuous self-assembly
To augment the previous yield data, we cloned and obtained nucleotide sequence data from RT-PCR products of gel-excised full-length molecules produced in self-assembly reactions resulting from single-site mutations at four of the possible six locations: W*, X*, Y* and *Z. In other words, we examined the effects of mutations at the 3′ ends of W, X and Y, plus in the h region of Z. For W* we tested only CAG mutants, but for the other three locations we tested all four mutants, for a total of 13 sets of sequence data. With data from 3–13 clones from each system (total = 81 clones; mean = 6.2 clones/system), we tabulated the existence of insertions at each junction (Table 2), and noted sequence identities (Figure 6) and any irregularities outside of the junctions.
Table 2.
Frequencies of tF2 reaction at each junction
| Location | Mutation | Non W·X·Y·Z | tF2 frequency in W·X·Y·Z clonesa | ||
|---|---|---|---|---|---|
| frequency | W–X junction | X–Y junction | Y–Z junction | ||
| W* | CAG | 0/13 | 11/13 | 13/13 | 4/13 |
| *X | None tested | – | – | – | – |
| X* | CAC | 1/6 | 0/5 | 5/5 | 1/5 |
| CAG | 2/6 | 0/4 | 4/4 | 1/4 | |
| CUU | 1/6 | 1/5 | 4/5 | 3/5 | |
| GAU | 0/6 | 4/6 | 6/6 | 4/6 | |
| *Y | None tested | – | – | – | – |
| Y* | CAC | 1/6 | 2/5 | 5/5 | 3/5 |
| CAG | 1/5 | 1/4 | 4/4 | 3/4 | |
| CUU | 0/6 | 2/6 | 6/6 | 1/6 | |
| GAU | 3/3 | – | – | – | |
| *Z | CAC | 0/6 | 0/6 | 6/6 | 3/6 |
| CAG | 0/6 | 4/6 | 6/6 | 5/6 | |
| CUU | 0/6 | 2/6 | 6/6 | 6/6 | |
| GAU | 1/6 | 1/5 | 5/5 | 5/5 | |
| All mutants: | 38% | 99% | 52% | ||
| Mutants with a mutation at this junction: | 84% | 95% | 68% | ||
| Mutants without mutation at this junction: | 29% | 100% | 33% | ||
| All CAU self-assembly (data from Table 1): | 25% | 100% | 40% | ||
atF2 mechanism assumed if an insertion of one or more nt exists at the junction.
Overall, the data followed the general trends of the sequences from the all-CAU system, but the deviations gave insight into how promiscuous self-assembly is possible. We first noted the appearance of a few clones that were the consequence of improper self-assemblies, which were not seen in the all-CAU system. Of the 81 clones, 10 were constructs such as W·Z·Z·Z and W·X·X·Z, each of which we observed three times. Most striking was the fact that the GAU systems were particularly prone to mis-assemble; when we engineered GAU into the 3′ end of Y (Y*), we only retrieved three clones and none of them was W·X·Y·Z. This corroborates our above finding that mutations in the 5′ position of CAU are the most damaging to the self-assembly process.
We then examined the junctions at the remaining 71 clones that did self-assemble properly. As expected, the X–Y junction was the most susceptible to insertions, although we did actually observe one clone, from CUU at X*, that displayed no insertion at this junction, implying that the R2F2 mechanism was used there. Also, in the other X–Y junctions we examined, the variability of insertion sequence lengths and identities was greater than that of the all-CAU system (Figure 6). At the W–Y and Y–Z junctions, insertion frequencies were somewhat higher than in the all-CAU system (Table 2). Importantly, there appears to be a correlation between whether a mutation was made at a particular junction and whether an insertion was present at that junction. When ‘non-mutated’ junctions were considered (i.e. there was a CAU present on both sides of the junction), the insertion frequencies matched those of the all-CAU system closely (Table 2). However, when either the CAU at the 3′ end of the 5′ fragment or at the 5′ end of the 3′ fragment was mutated, the tF2 mechanism was invoked with greater frequency. For instance, at the W–X junction, mutations at W* or *X raise the pinch rate from 25–29% to 84%. This can be seen visually; when mutations were added at *X, the W·X intermediate increases in size (Figure 5, compare lanes 1 and 2 with 3 and 4). This shows that the R2F2 mechanism is more inhibited by mutations in the IGS complement, a consequence perhaps of a need for two IGS–IGS complement interactions in R2F2 but only one in tF2.
Lastly, by examining the effects of mutations when placed in the h region, for which we only tested *Z, we were able to confirm the mechanism outlined in Figure 1A. All four of the mutant triplets tested, CAC, CAG, CUU and GAU, when engineered into the h region of Z, result in an insertion that includes that particular mutation when the tF2 mechanism is employed. For example, 5 of 6 clones examined for self-assembly with CAG at *Z exhibited insertions at the Y–Z junction, and all of these insertions included the triplet CAG. Across all *Z assays using mutants, no insertions at Y–Z contained CAU. Conversely, the mutant triplet never appeared at other junctions. Thus a portion of the h region is in fact the source of the insertion, although the exact location of the cross-strand attack is somewhat variable, especially when the IGS complement is mutated.
Azoarcus ribozyme mis-cleavage assays
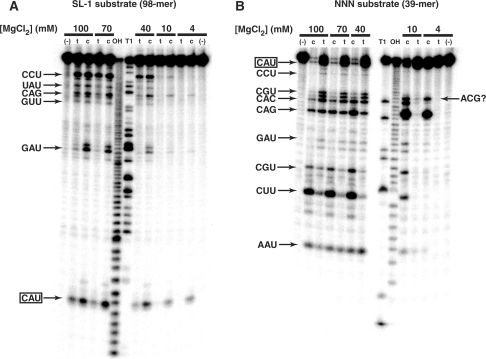
The obvious means by which mutations in the IGS complement can be tolerated during self-assembly is through promiscuous mis-pairing with the IGS, a situation that can be exacerbated by high ionic strengths and lower temperatures. Thus we sought to test the ability of the wild-type Azoarcus ribozyme to bind to—and effect catalysis at—sites other than CAU in a non-self-assembly setting. We used two 5′-end-labled substrate RNA targets and assayed the ability of both the covalent Azoacrus ribozyme and the non-covalent four-piece trans complex to cleave these substrates following CAU and other triplets. One target was the 98-mer SL-1, which represents one-half of the class I ligase ribozyme (17). It contains only one CAU triplet, and canonical cleavage by Azoarcus ribozyme should target that site only, and generate a 5-mer and a 93-mer. Several other CAU-like triplets exist in this molecule by chance. The second was a 39-mer construct NNN, which we designed to contain exactly one CAU triplet and at least one copy of each possible variant in which one of the three nucleotides is mutated. We performed all assays at 48°C and in 4–100 mM MgCl2 for 30 min using 2 μM of each RNA species.
Both target substrates demonstrated that the Azoarcus ribozyme can mis-cleave under these conditions (Figure 7). For the SL-1 substrate, a strong cleavage band 5 nt in size corresponding to the CAU site could be seen at all MgCl2 concentrations (Figure 7A). However, as the [MgCl2] was raised to 40 mM and above, cleavage could be seen at other triplets, especially CCU, CAG and GAU. For the NNN substrate, a similar pattern appeared (Figure 7B). Cleavage at CAU was seen at the lowest [MgCl2] values, but CAG was clearly a target under those conditions as well. As [MgCl2] was raised, CAC, CGU, CUU and CCU began to become occasional targets. Cleavage at other sites was always weak at best, with the exception of an aberrant scission after an ACG triplet. Note that quantification of cleavage is not readily feasible using 5′-end-labeled substrates because multiple cleavage events cause bands to appear and then disappear; the ‘loss’ of the CAG cleavage product at the highest [MgCl2] values is a good example (Figure 7B). Nevertheless, we made a qualitative ranking of Azoarcus ribozyme-directed triplet cleavage preferences in these two substrates: CAU > CAG > CAC > CGU = CUU > CCU > GAU = AAU = GUU > all others. This ordinal ranking complements the notion that mutations are increasingly tolerated as one proceeds 5′ to 3′ in the IGS complement triplet. We also observed that the four-piece trans complex displayed the same preferences for triplet cleavage, although in general higher [MgCl2] values are required to support catalysis, as expected.
Figure 7.
Mis-cleavage of target substrates by Azoarcus catalysts. (A) Cleavage of 5′-end-labeled substrate SL-1 by wild-type L8 covalently contiguous Azoarcus ribozyme (c) and by a trans complex formed by an equimolar mixture of W, h·X, h·Y and h·Z oligomers (t). Incubation time is 20 min, which does not allow enough time for significant oligomer self-assembly into covalent ribozymes to occur. Triplets immediately 5′ of major cleavage sites are indicated. As [MgCl2] is raised, more cleavage products are seen, and larger products get cleaved into smaller products by multiple endonuclease events. T1 and OH− ladders, possessing 3′ cyclic phosphate moieties, run slightly faster than Azoarcus-cleaved RNAs, which possess 3′-OH moieties. First and last lanes, no catalyst added. (B) Cleavage of the 5′-end-labeled substrate NNN, which has at least one copy of all triplets that are one point mutation away from CAU. Triplets immediately 5′ of major cleavage sites are indicated. Cleavage after triplet ACG is unexplained. First and last lanes, no catalyst added.
DISCUSSION
We have demonstrated that the canonical IGS complement triplet CAU of the Azoarcus ribozyme is not strictly required for the self-construction of this RNA from component fragments. Mutations at the 3′ uridine of this triplet are particularly tolerated by the system, although alterations of the C and the A can be made and self-assembly will be retained. All mutations, however, lower the yield of self-assembly, presumably because the system is then forced to operate with a mismatch between the IGS and its complement. Such mis-pairing seems only possible at abnormally high ionic strength conditions such as 100 mM MgCl2, conditions that also are favorable for the formation of multipartate trans complexes.
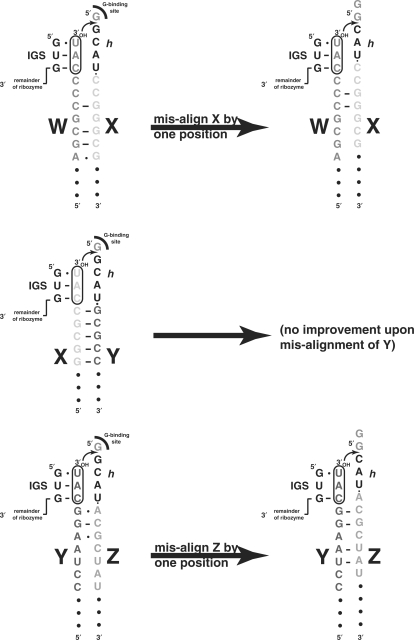
Deviations away from CAU in the oligonucleotides also appear to favor the tF2 mechanism over the R2F2 mechanism. This is most apparent at the W–X and Y–Z junctions, where the all-CAU system tends to foster the R2F2 mechanism as we originally outlined it (12). Because of the exact placement of the CAU sequence, the X–Y junction turned out to be predisposed to accommodate the tF2 mechanism (12), but it is possible to draw oligomer alignments at all three junctions that would allow the single-step tF2 reaction to take place (Figure 8, left). The W and h·X oligomers can be aligned with geometries analogous to those of X and h·Y, but with only three local Watson–Crick pairs and one A–G wobble pair. The Y and h·Z oligomers can be similarly aligned but with only one Watson–Crick pair and two A–G wobbles. Of note is that the Y and h·Z base pairs, though weak, occur adjacent to the CAUs themselves, while the X and h·Y base pairs are two positions away. This could explain why we observed a higher frequency of tF2 reactions at the Y–Z junction, in both the all-CAU and the mutant systems.
Figure 8.
Possible oligomer alignments involved in the tF2 mechanism. On the left are analogous alignments between adjacent oligomers that maintain the spatial arrangements of the IGS and the guanosine-binding sites in the catalytic pocket of the ribozyme. This alignment generates the most commonly observed insertion, GCAU. The conceivable base pairing that stabilizes the substrate oligomers is shown, with relative strengths near the CAU triplets being X–Y >> Y–Z > W–X, in accordance with the observed tF2 frequencies at these junctions. On the right are conceivable mis-alignments for the W–X and Y–Z junctions that lead to the next most frequently observed insertion, CAU.
From our insertion frequency data (Tables 1 and 2) we infer that weakening of the IGS–IGS complement interaction hinders self-assembly mainly by inhibiting the chemical step of splicing, thereby favoring reaction pathways reliant on fewer steps, such as tF2 when compared to R2F2 (Figure 1). The rates of both the Tetrahymena and the Anabaena group I intron-catalyzed self-splicing reactions can be affected by mismatches in the P1 stem that encompasses the IGS. In the case of the Tetrahymena rRNA intron, mismatches in the 6 nt IGS actually can enhance reaction rates in vitro because product release is rate limiting (18). But in the cases of the Anabaena and Azoarcus ribozymes, both derived from tRNA introns with 3 nt IGS regions, product release is not limiting and the chemical step becomes more kinetically dominant (11,16). It is of some interest to note the analogy between the 5′ nt of the IGS complement in these ribozymes and the 5′ ‘wobble’ position of the tRNA anticodon. Watson–Crick pairing at the other two positions in the triplet seems to be more critical in correct guiding operations.
Promiscuous self-assembly, that is, when mutations are made in one or more CAU triplets in either the h regions or at the 3′ ends of oligomers, must be manifest through either a mis-cleavage event or, in the case of the tF2 mechanism, a mis-alignment event. We show here that the Azoarcus ribozyme can mis-cleave at relatively high MgCl2 concentrations and relatively low temperatures (Figure 7). When CAU is mutated, proper alignment with the IGS and phosphotransfer catalysis would require that at least one base-pairing interaction be weakened. Crystal structures of the Azoarcus ribozyme trapped at various stages of the splicing cascade reveal a very structured environment at the active site with two key Mg(II) ions contributing to structural stability (19,20). The wobble-pair interaction at the splice site between G10 of the intron (the 5′ G of the ribozyme here) and U(–1) of the exon (the 3′ U of the CAU here) specifically involves an H-bonding network that includes other residues in the ribozyme. Even the least deleterious mutation, CAC, would disrupt this evolved network; a cis Watson–Crick/Watson–Crick G–C pair at the 5′ end of the IGS would not be possible without structural perturbation, because both protons on the extra-cyclic amine of G10 are involved in donating H-bonds to another residue, A87 (19). In the case of tF2 mechanism, in addition to mismatches between the IGS and its complement that hinder, but do not completely retard, catalysis, alternative alignments of the duplex substrate can presumably facilitate cross-strand attack (Figure 8, right), and supporting this notion is the variability of insertion sizes (1–5 nt) that can be seen at all three junctions. Nevertheless, the GCAU insertion is always the most common, as the geometry that generates a 4 nt insertion probably preserves the relative locations of the IGS and the guanosine-binding site of the ribozyme.
These results confirm that RNA catalysis, and ribozyme self-assembly in particular, have fewer constraints under permissive environmental conditions. A model in which catalysts spontaneously coalesce from a pool of random oligomers generated from an abiotic source would certainly require a high degree of tolerance for sequence variability. Many positions in real ribozymes have been shown to be mutable, such that the fitness landscape for catalytic function is not prohibitively rugged (21). We have provided evidence here that even one of the most essential elements of a recombinase ribozyme, the IGS–IGS complement pair, can withstand some mutations away from an optimal sequence. Mutations in the remainder of the oligomers should also be tolerable, and it will be of great interest to observe the selection in vitro of self-assembling autocatalytic sets from randomized populations. Although this scenario is likely dependent on the stabilizing influence of high salt, some have proposed prebiotic synthesis reactions in the high salinity that characterized Earth's early oceans [e.g. (22,23); however, see Ref. (24)]. Given that the tF2 mechanism requires fewer steps, we propose that a recombination-based self-assembly of genetic information would have begun with that mechanism, and later was superseded by the R2F2 mechanism that allowed a distillation of the self-recognition tag (14) down to as few as 3 nt.
ACKNOWLEDGEMENTS
We thank Lisa Thomas for technical assistance. This project was supported by a research grant from the National Aeronautics and Space Administration (NNG04-GM20G) to N.L. Funding to pay the Open Access publication charges for this article was provided by NASA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lohrmann R, Bridson PK, Orgel LE. Efficient metal-ion catalyzed template-directed oligonucleotide synthesis. Science. 1980;208:1464–1465. doi: 10.1126/science.6247762. [DOI] [PubMed] [Google Scholar]
- 2.Inoue T, Orgel LE. A nonezymatic RNA polymerase model. Science. 1982;219:859–862. doi: 10.1126/science.6186026. [DOI] [PubMed] [Google Scholar]
- 3.Ferris JP, Hill A.R., Jr, Liu R, Orgel LE. Synthesis of long prebiotic oligomers on mineral surfaces. Nature. 1996;381:59–61. doi: 10.1038/381059a0. [DOI] [PubMed] [Google Scholar]
- 4.Chetverin AB, Chetverina HV, Demidenko AA, Ugarov VI. Nonhomologous RNA recombination in a cell-free system: evidence for a transesterification mechanism guided by secondary structure. Cell. 1997;88:503–513. doi: 10.1016/S0092-8674(00)81890-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chetverina HV, Demidenko AA, Ugarov VI, Chetverin AB. Spontaneous rearrangements in RNA sequences. FEBS Lett. 1999;450:89–94. doi: 10.1016/s0014-5793(99)00469-x. [DOI] [PubMed] [Google Scholar]
- 6.Lutay AV, Zenkova MA, Vlassov VV. Nonenzymatic recombination of RNA: possible mechanism for the formation of novel sequences. Chem. Biodivers. 2007;4:762–767. doi: 10.1002/cbdv.200790062. [DOI] [PubMed] [Google Scholar]
- 7.Zaug AJ, Cech TR. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986;231:470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- 8.Doudna JA, Szostak JW. RNA-catalysed synthesis of complementary-strand RNA. Nature. 1989;339:519–522. doi: 10.1038/339519a0. [DOI] [PubMed] [Google Scholar]
- 9.Mörl M, Schmelzer C. Group II intron RNA-catalyzed recombination of RNA in vitro. Nucleic Acids Res. 1990;18:6545–6551. doi: 10.1093/nar/18.22.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riley CA, Lehman N. Generalized RNA-directed recombination of RNA. Chem. Biol. 2003;10:1233–1243. doi: 10.1016/j.chembiol.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Zaug AJ, McEvoy MM, Cech TR. Self-splicing of the group I intron from Anabaena pre-tRNA: requirement for base-pairing of the exons in the anticodon stem. Biochemistry. 1993;32:7946–7953. doi: 10.1021/bi00082a016. [DOI] [PubMed] [Google Scholar]
- 12.Hayden EJ, Lehman N. Self-assembly of a group I intron from inactive oligonucleotide fragments. Chem. Biol. 2006;13:909–918. doi: 10.1016/j.chembiol.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 13.Kauffman SA. The Origins of Order. New York, NY: Oxford University Press; 1993. [Google Scholar]
- 14.Levy M, Ellington AD. The descent of polymerization. Nat. Struct. Biol. 2001;8:580–582. doi: 10.1038/89601. [DOI] [PubMed] [Google Scholar]
- 15.Burton AS, Lehman N. Calcium(II)-dependent catalytic activity of the Azoarcus ribozyme: testing the limits of resolution for in vitro selection. Biochimie. 2006;88:819–825. doi: 10.1016/j.biochi.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Kuo LY, Davidson LA, Pico S. Characterization of the Azoarcus ribozyme: tight binding to guanosine and substrate by an unusually small group I ribozyme. Biochim. Biophys. Acta. 1999;1489:281–292. doi: 10.1016/s0167-4781(99)00200-6. [DOI] [PubMed] [Google Scholar]
- 17.Hayden EJ, Riley CA, Burton AS, Lehman N. RNA-directed construction of structurally complex and active ligase ribozymes through recombination. RNA. 2005;11:1678–1687. doi: 10.1261/rna.2125305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 2. Kinetic description of the reaction of an RNA substrate that forms a mismatch at the active site. Biochemistry. 1990;29:10172–10180. doi: 10.1021/bi00496a004. [DOI] [PubMed] [Google Scholar]
- 19.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 20.Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- 21.Kun Á., Santos M, Szathmáry E. Real ribozymes suggest a relaxed error threshold. Nat. Genet. 2005;37:1008–1011. doi: 10.1038/ng1621. [DOI] [PubMed] [Google Scholar]
- 22.Lathe R. Fast tidal cycling and the origin of life. Icarus. 2004;168:18–22. [Google Scholar]
- 23.Baaske P, Weinert FM, Duhr S, Lemke KH, Russell MJ, Braun D. Extreme accumulation of nucleotides in simulated hydrothermal pore systems. Proc. Natl Acad. Sci. USA. 2007;104:9346–9351. doi: 10.1073/pnas.0609592104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monnard P.-A, Apel CL, Kanavarioti A, Deamer DW. Influence of ionic inorganic solutes on self-assembly and polymerization processes related to early forms of life: implication for a prebiotic aqueous medium. Astrobiology. 2002;2:139–152. doi: 10.1089/15311070260192237. [DOI] [PubMed] [Google Scholar]