Abstract
To identify genes involved in poly(A) metabolism, we screened the yeast gene deletion collection for growth defects in the presence of cordycepin (3′-deoxyadenosine), a precursor to the RNA chain terminating ATP analog cordycepin triphosphate. Δpho80 and Δpho85 strains, which have a constitutively active phosphate-response pathway, were identified as cordycepin hypersensitive. We show that inorganic polyphosphate (poly P) accumulated in these strains and that poly P is a potent inhibitor of poly(A) polymerase activity in vitro. Binding analyses of poly P and yeast Pap1p revealed an interaction with a kD in the low nanomolar range. Poly P also bound mammalian poly(A) polymerase, however, with a 10-fold higher kD compared to yeast Pap1p. Genetic tests with double mutants of Δpho80 and other genes involved in phosphate homeostasis and poly P accumulation suggest that poly P contributed to cordycepin hypersensitivity. Synergistic inhibition of mRNA synthesis through poly P-mediated inhibition of Pap1p and through cordycepin-mediated RNA chain termination may thus account for hypersensitive growth of Δpho80 and Δpho85 strains in the presence of the chain terminator. Consistent with this, a mutation in the 3′-end formation component rna14 was synthetic lethal in combination with Δpho80. Based on these observations, we suggest that binding of poly P to poly(A) polymerase negatively regulates its activity.
INTRODUCTION
Inorganic polyphosphate (poly P) comprises chains of 10s to 100s of phosphate residues, connected by energy-rich phospho-anhydride bonds. Despite the fact that poly P is an ubiquitous molecule detected in every living cell, our knowledge concerning its biochemistry and biological function remains incomplete (1,2). Poly P serves important roles as a substitute for ATP for sugar and adenylate kinases (3,4), as phosphate reservoir with osmotic advantage (5–8), as energy source (9) and reservoir (10), as buffer against alkaline stress (11) and as chelator of divalent ions (1).
In addition to these general functions, poly P has been implicated in a number of regulatory processes both in prokaryotes and eukaryotes. In Escherichia coli, poly P is essential for survival during stationary phase (12). It accumulates into large amounts in response to stress situations brought about by nutritional downshift from rich to minimal media (13), phosphate limitation (14), amino acid depletion (15) or nitrogen starvation. During the latter condition, poly P promotes ribosomal protein degradation by binding and activating the Lon protease in E. coli delivering amino acids needed to respond to starvation (16). Poly P is also ubiquitous in mammalian cells and tissues (17) where it has been implicated in a host of regulatory processes. For example, the mammalian TOR kinase, which is involved in cell growth and proliferation (18), is activated by poly P under conditions of nutritional starvation (19). This observation also led to the proposal of an evolutionary conserved role for poly P in stress response (19). Moreover, poly P was found to enhance proliferation of human fibroblast cells (20), to stimulate calcification of osteoblast-like cells (21), to inhibit the secretion of immunoglobulin and to stimulate apoptosis in human plasma and myeloma cells (22), and more recently, poly P has been shown to have anti-metastatic and anti-angiogenic activities (23).
Interestingly, there are several reports that link poly P to the regulation of gene expression through effects on RNA polymerase transcription. In E. coli, poly P is associated with RNA polymerase during the stationary phase and inhibits specifically the transcriptional activity of the enzymes associated with the Eσ70 promoter-recognition subunit, which is involved in the transcription of genes during exponential growth phase (24). Moreover, poly P induces transcription of rpoS, the stationary phase σ factor (25). Polyphosphate kinase (Ppk1), the enzyme synthesizing poly P, is a component of the E. coli RNA degradosome (26). Since poly P is a potent inhibitor of the degradosome, it was suggested that Ppk1 might act to maintain a correct microenvironment for proper mRNA degradation (26). In addition, poly P has been shown to associate with ribosomes and to suppress misincorporation of amino acids during translation (27).
In yeast, poly P concentrations can reach ∼120 mM (1) and thus poly P contributes up to 20% of the cellular dry weight. Most of the poly P (90–99%) is localized to the vacuole (28,29), but poly P was also detected in the cytoplasm and the nuclei of yeast (30,33). For rat liver nuclei, micromolar poly P concentrations were reported (17) and for yeast nuclei a similar level of poly P is assumed (1). Poly P content of a yeast cell is strongly dependent on the growth phase (31,32) and the average length of nuclear poly P polymers of approximately 45 phosphate residues was found to be changing dynamically with growth conditions (33). Enzymes involved in eukaryotic poly P synthesis remain mostly elusive to this day (1), but activities indicative of an active poly P catabolism have been associated with all cellular compartments (31,34), including the nucleus (30,35). The observation that a double mutant of PPN1 [encoding a endopolyphosphatase; (36)] and PPX1 [encoding an exopolyphosphatase; (37)] rapidly looses viability in stationary phase (36), underscores the biological importance of poly P. We recently screened the entire collection of haploid yeast gene deletion mutants for poly P content (38) and found that poly P metabolism and primary metabolism (e.g. ATP and phosphate homeostasis) were strongly interdependent. This screen also revealed that all cellular compartments are linked to poly P homeostasis (38).
In this work, we establish a connection between cellular accumulation of poly P and inhibition of poly(A) polymerase activity. Yeast strains mutant in central components of the phosphate response pathway (Δpho80 and Δpho85) contained increased amounts of poly P that contributed to cordycepin-hypersensitive growth. We propose a role for poly P in negatively regulating polyadenylation through inhibition of poly(A) polymerase.
MATERIALS AND METHODS
Yeast strains and plasmids
Wild-type BY4741 (Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) and isogenic mutant strains were obtained from EUROSCARF. The rna14-1 genotype is Mat a; ura3-1; trp1-1; ade2-1; leu2-3; 112; his3-11; 15; rna14-1 (39). Double mutants were generated by disrupting the PHO80 open reading frame with a NatR cassette by homologous recombination in Δpho4, Δpho2, Δpho84 and rna14-1 strains. PHO80 and PHO85 genes were cloned into pRS313 using BamHI and NotI restriction sites following PCR amplification from genomic DNA with primers PHO80-5′ AAGATCGGATCCCTTTCTATGGAAATATGAATG and PHO80-3′ GATCTTGCGGCCGCAAAGAACAGTGATGATATGAAT and PHO85-5′ AAGATCGGATCCTGTTTTAGAAATATGTGCACT and PHO85-3′ GATCTTGCGGCCGCTTTACGTTCTGCTCTCTCACTT. Yeast strains were grown at 30°C either in YPD (1% yeast extract, 2% tryptone, 2% glucose) or in synthetic complete medium (SD, yeast nitrogen base and complete amino acid mixture plus 2% glucose). SD was supplemented with cordycepin (40 μg/ml) purchased from Sigma or 5′-Fluoroorotic acid (1 mg/ml, Zymo Research, Orange, CA, USA).
Poly(A) length analysis
Assays were performed essentially as described (40). In the standard reaction, 2 μg of total RNA was incubated with 400 ng recombinantly expressed yeast poly(A) polymerase (a gift from G. Martin, Basel) and 0.2 μl [α-32P]-cordycepin triphosphate (Perkin–Elmer) in reaction buffer (20 mM Tris–HCl pH 7.0, 50 mM KCl, 0.7 mM MnCl2, 10% Glycerol, 100 μg/ml BSA) for 30 min at 30°C in a total volume of 12 μl. After heat inactivation, RNA was digested with RNase A and RNase T1, followed by proteinase K treatment. Precipitated RNAs were resolved on 15%/8.3 M urea polyacrylamide gels, that were exposed and visualized on a FLA-7000 phosphor-imager (Fuji).
Polyadenylation assays
Conditions were as described in Refs (41) and (42) with modifications. The yeast poly(A) polymerase reaction mixture of 15 μl contained 100 ng yeast poly(A) polymerase (a generous gift from G. Martin, Basel), 5 pmol of 5′-end labeled A15 RNA primer, 0.5 mM ATP, 5 mM MgCl2, 25 mM Tris–HCl pH 7.9, 20 mM KCl, 10% glycerol, 0.01 mM EDTA, 0.1 mg/ml BSA, 1 mM DTT and 0.02% Nonidet P-40. The reaction mixture with bovine poly(A) polymerase consisted of 200 ng bovine poly(A) polymerase (a gift from G. Martin, Basel), 5 pmol of 5′ end-labeled A15 RNA primer, 0.5 mM ATP, 25 mM Tris–HCl pH 8.3, 40 mM KCl, 6 mM MgCl2, 0.05 mM EDTA, 0.5 mM DTT, 0.01% Nonidet P-40, 10% glycerol and 200 μg/ml BSA in a total volume of 15 μl. Poly P was added to the reactions as indicated in the figure legends. Reactions were incubated at 30°C for the indicated time and stopped by addition of 25 mM EDTA. Reaction products were precipitated and resolved on 15%/8.3 M urea polyacrylamide gels that were exposed and visualized on a FLA-7000 phosphor-imager (Fuji).
Purification of recombinant E. coli polyphosphate kinase
The gene encoding the E. coli polyphosphate kinase [EcPPK; (43)] was amplified with the primers CATGCCATGGGTCAGGAAAAGCTATACATCG and CGCGGATCCTGCGGACGAGGGGATTTATCG and cloned with the NcoI and BamHI sites (underlined) into the expression vector pETM41 (EMBL Protein Expression and Purification Unit). EcPPK was expressed as a fusion with a maltose-binding protein in E. coli BL21 cells and purified on amylose resin following standard protocols (New England BioLabs, Beverly, MA, USA). The activity was determined with the reverse reaction by measuring ATP synthesis at 37°C in 50 μl reactions containing 10 μM poly P88, 1 μM ADP, 50 mM Tris and 50 mM malate, pH 6.5 and purified EcPPK. The reactions were stopped by heat inactivation at 90°C for 2 min. ATP was quantified after the addition of 50 μl luciferase reaction mixture (ATP bioluminescence assay kit CLS II, Roche Molecular Biochemicals) in a luminometer [Lumat LB 9507 (Berthold Technologies GmbH & Co. KG)].
Poly P measurements and poly P synthesis in vitro
Determination of poly P concentration was performed as previously described (32). Poly P with an average chain length of 750 residues was synthesized as described (13) except for some modifications. Poly P was synthesized in 200 μl reactions containing 138 nM [γ-32P]-ATP, 1 mM ATP, 2 mM creatine-phosphate, 6 U creatine phosphokinase (from rabbit muscle, Sigma-Aldrich), 150 U polyphosphate kinase (1 unit corresponds to the transfer of 1 pmol Pi to ADP/min), 5 mM MgCl2, 50 mM Tris and 50 mM malate (pH 6.5). The reactions were incubated for 16 h at 30°C. 32P-labeled poly P was purified as described (33) and was eluted in 100 μl water. Radioactivity was measured in a scintillation counter (LS1801, Beckman Instruments, Fullerton, California) and the poly P concentration was calculated based on the fraction of 32P that was incorporated in poly P (resulting typically in ∼3 mM considering phosphate residues).
Poly P binding
Poly P binding assays were performed as reported earlier (16). Purified poly(A) polymerase from Saccharomyces cerevisiae and Bos taurus was diluted to a concentration of 200 nM in 100 μl reaction buffer (50 mM Tris–HCl pH 7.5, 5 mM MgCl2). Equal volumes of radioactive poly P750 (0.3–30 nM, 890 c.p.m./pmol for yeast and 132 000 c.p.m./pmol for bovine poly(A) polymerase) were added. After 5 min at 37°C, the mixtures were applied to nitrocellulose filter discs (0.45 μm) and washed twice with 1 ml ice cold TBS (50 mM Tris–HCl pH 7.5, 100 mM NaCl). The remaining radioactivity on the filters, corresponding to the amount of poly P–protein complex, was measured in a scintillation counter.
RESULTS
Δpho85 and Δpho80 strains are hypersensitive towards cordycepin
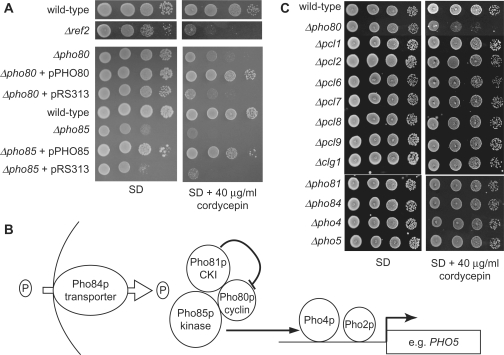
Poly(A) addition by poly(A) polymerase is terminated in vitro in the presence of cordycepin triphosphate (CoTP) (44,45). Since temperature-sensitive mutations in the essential 3′ end formation factors Rna14p and Rna15p render yeast cells sensitive to cordycepin (3′-deoxyadenosine) (46), we reasoned that a growth phenotype in the presence of this drug may identify mutant strains involved in poly(A) metabolism. Cordycepin is taken up by the yeast (47) and converted into the RNA-chain terminating CoTP, which is a substrate for RNA synthesis. The presence of 40 μg/ml cordycepin in the medium has only mild toxic effects on wild-type cells and increased the doubling time in liquid culture by ∼20% (data not shown). In contrast, a strain lacking the 3′ end formation factor Ref2p (48) showed severe growth inhibition (Figure 1A). Thus, strains lacking non-essential genes involved in poly(A) metabolism can be identified through cordycepin-hypersensitive growth.
Figure 1.
Δpho85 and Δpho80 strains display cordycepin-hypersensitive growth. (A) Twenty-fold serial dilutions of the indicated strains were spotted on agar plates that either lacked (SD) or contained 40 μg/ml cordycepin; plates were photographed after 72 h of incubation at 30°C. Complementation of Δpho80 and Δpho85 phenotypes was tested with empty plasmid (pRS313) or with the same vector carrying the PHO80 or PHO85 genes, respectively. (B) Schematic representation of key components of the PHO pathway (50). Pho84p is a high-affinity phosphate transporter, Pho85p and Pho80p form a cyclin-CDK (cyclin-dependent kinase) pair and Pho81p is an associated CDK inhibitor; the Pho4p transcription factor (the target of the Pho85p/Pho80p complex) in its hypo-phosphorylated form associates with the homeodomain transcription factor Pho2p to drive the expression of phosphate-regulated genes, like e.g. the Pho5p secreted acid phosphatase. (C) Cordycepin hypersensitivity is restricted to PHO pathway-dependent functions of Pho85p. Drop test as in A) with mutant yeast strains lacking the indicated Pho85p cyclins (upper panel) and components of the PHO pathway (lower panel) on medium lacking or containing 40 μg/ml cordycepin.
We screened a haploid S. cerevisiae gene deletion collection (49) for growth defects in the presence of cordycepin using a simple drop test on agar plates. The entire results of this screening will be presented elsewhere (S.H. and B.D., unpublished data). Here we focus on the further characterization of cordycepin-hypersensitive growth observed with Δpho85 and Δpho80 strains (Figure 1A). Complementation of the mutant strains with plasmids carrying wild-type PHO85 and PHO80 genes, respectively, demonstrated a direct requirement for these genes for cordycepin resistance. Empty plasmids, in contrast, did not complement. Pho85p kinase and its cyclin partner Pho80p form a complex and are central regulators of the PHO pathway in yeast (Figure 1B) (50). However, Pho85p can associate with nine other cyclins to perform functions in various cellular pathways including the cell cycle, carbon-source utilization and glycogen metabolism (51). We also tested mutant strains of seven other Pho85p cyclins (Figure 1C; Δpcl1, Δpcl2, Δpcl6, Δpcl7, Δpcl8, Δpcl9 and Δclg1). Since we could not observe sensitivity with these strains, the cordycepin-hypersensitive growth of Δpho85 and Δpho80 appeared to be mainly linked to their function in phosphate homeostasis. The absence of Pho85p or Pho80p results in constitutively active transcription of phosphate-dependent genes (50). To evaluate the relation of other PHO pathway components and the observed cordycepin sensitivity, we analyzed strains lacking the transcription factors Pho4p and Pho2p, the cyclin-dependent kinase inhibitor Pho81p, the high-affinity phosphate transporter Pho84p and the target gene PHO5 (encoding a secreted acid phosphatase). None of these strains reacted to the drug (Figure 1C and data not shown). We conclude that a constitutively active PHO pathway underlies the observed cordycepin hypersensitivity of Δpho85 and Δpho80 mutants.
Δpho85 and Δpho80 strains accumulate an inhibitor of poly(A) polymerase
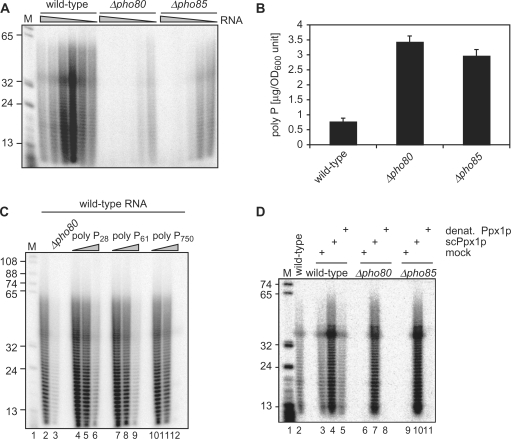
Since CoTP can act as a terminator of the poly(A) addition reaction in vitro (44,45), we considered the possibility that cordycepin hypersensitivity of Δpho85 and Δpho80 strains may relate to this process. Therefore, we isolated total RNA from wild-type and mutant strains and analyzed the length distribution of cellular poly(A). The protocol for poly(A) labeling that we employed included the transfer of radioactively labeled [α32P]-CoTP to 3′-OH groups present in the RNA preparation with recombinant yeast poly(A) polymerase (40). With wild-type RNA, a uniformly distributed length of approximately 10–70 adenosines was observed (Figure 2A). Quite unexpectedly, we were unable to label any poly(A) isolated from Δpho85 and Δpho80 strains under standard assay conditions (i.e. 1–2 μg of total RNA; Figure 2A). We considered the possibility that some inhibitory activity may be present in the RNA preparations of the mutant strains interfering with the activity of poly(A) polymerase and performed the labeling assay with decreasing amounts of total RNA. We observed that a 4- to 8-fold dilution of the total RNA from Δpho85 and Δpho80 strains indeed allowed poly(A) labeling (Figure 2A). Furthermore, reduced amounts of wild-type RNA gave increased labeling efficiency. We conclude that an inhibitory activity was present in the total RNA preparations from Δpho85 and Δpho80 strains interfering with the poly(A) labeling test and that the same inhibitor was also present in wild-type RNA, albeit at lower concentration.
Figure 2.
Δpho85 and Δpho80 strains accumulate poly P, an inhibitor of poly(A) polymerase. (A) Poly(A)-labeling assays with total RNA extracted from wild-type, Δpho80 and Δpho85 strains. Decreasing amounts of total RNA (2 μg, 1 μg, 0.5 μg, 250 ng, 125 ng and 75 ng) were included with recombinant yeast Pap1p in the reactions. Following RNase A/T1 digestion, the labeled poly(A) tails were loaded on a 15% denaturing polyacrylamide gel. Poly(A) labeling was observed in Δpho80 and Δpho85 only following dilution of the RNA included in the reactions. HpaI-digested pBR322 fragments were 5′-end labeled and served as marker bands (M) with the indicated size. (B) Poly P contents in wild-type, Δpho80 and Δpho85 strains following growth in YPD medium for 6 h. (C) Poly(A) labeling assay as described in (A) with the addition of Δpho80 RNA (lane 3) or of increasing amounts of poly P (60 nM to 6 µM poly P28; 27 nM to 2.7 µM poly P61; and 2 nM to 200 nM poly P750) with the indicated average chain length (lanes 4–12) to poly(A)-labeling assays with wild-type RNA. Lane 2 shows the reaction of wild-type RNA without addition of inhibitors. (D) Poly(A)-labeling assay as described in (A) with total RNA obtained from wild-type, Δpho80 and Δpho85 strains. RNAs were treated as indicated with yeast exopolyphosphatase (scPpx1p), with heat-denatured exopolyphosphase (denat. Ppx1p) or buffer (mock) before the labeling reaction was performed.
Poly P accumulates in Δpho85 and Δpho80 strains and interferes with RNA 3′ end labeling by poly(A) polymerase
We hypothesized that the co-purification of a metabolite with Δpho85 and Δpho80 RNA may cause the inhibition of the poly(A) labeling reaction. It was suggested that the PHO pathway is involved in regulating the synthesis of poly P (6). However, inconsistent results were obtained when Δpho85 and Δpho80 strains were previously analyzed for poly P contents (6,52). Therefore, we measured poly P levels in wild-type and mutant strains with a recently developed assay (32) and found that Δpho85 and Δpho80 strains had approximately 3-fold more poly P than wild type (Figure 2B). In contrast, mutants in other components of the PHO pathway did not show this accumulation; indeed poly P concentrations were reduced in Δpho4, Δpho2 and Δpho84 mutant strains (Figure 5D) (6,53). These results support the idea that poly P metabolism is regulated by the PHO pathway and that constitutive activity of this pathway (in the absence of Pho85p and Pho80p) resulted in increased cellular poly P concentrations.
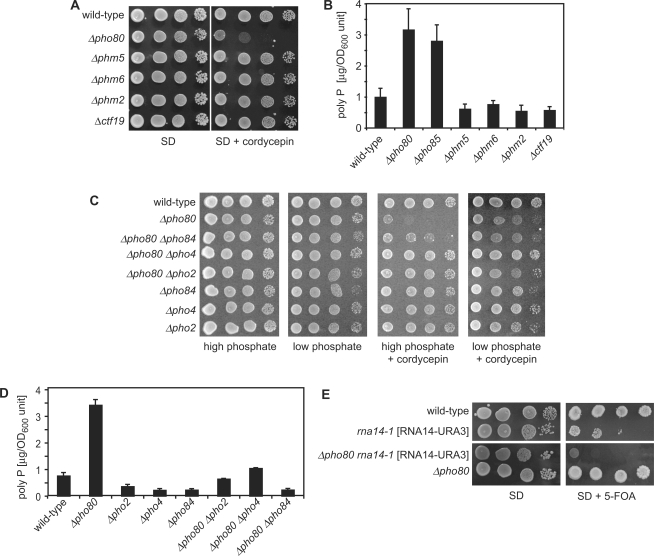
Figure 5.
Cellular poly P contents correlates with cordycepin-hypersensitive growth. (A) Drop test as described in Figure 1A with the indicated strains on agar plates containing or lacking cordycepin (40 μg/ml) as indicated. Plates were incubated for 72 h at 30°C. (B) Poly P content of the indicated strains was determined following growth in YPD medium for 6 h as described. (C) Drop test of 20-fold serial dilutions of the indicated strains on agar plates containing 10 mM (high phosphate) or 0.1 mM (low phosphate) phosphate and cordycepin (40 μg/ml) as indicated. Plates were incubated for 72 h at 30°C. (D) Poly P content of the indicated strains was determined following growth in YPD medium for 6 h. (E) The Δpho80 mutation is synthetic lethal in combination with rna14-1. The PHO80 gene was deleted in a rna14-1 strain that carried a plasmid with wild-type RNA14 and the URA3 marker gene. Serial dilutions of indicated wild-type, single and double mutant strains were spotted on synthetic complete medium lacking (SD) or containing the drug 5-Fluoroortic Acid (SD + 5-FOA) and incubated for 72 h at 30°C. Since 5-FOA selects against the URA3 marked plasmid, lack of growth in the presence of the drug indicated that cell viability was dependent on the wild-type copy of RNA14 present on the plasmid.
The accumulation of poly P in Δpho85 and Δpho80 strains prompted us to test whether poly P is an inhibitor of poly(A) polymerase. Therefore, we repeated the poly(A) labeling test with standard amounts of RNA (2 μg) and added either Δpho80 RNA or increasing amounts of poly P with different average chain length (21, 61 and 750 phosphate residues). We observed that both the addition of Δpho80 RNA and of poly P resulted in inhibition of the labeling reaction (Figure 2C). The inhibition was nearly complete with the highest concentration of poly P employed (200 nM). However, in multiple repetitions of this experiment, we could not observe a clear correlation of the degree of inhibition and the chain length of poly P included in the assay.
To demonstrate that poly P is indeed the inhibitory activity present in the Δpho85 and Δpho80 RNA preparations, we treated the RNAs with recombinant yeast exopolyphosphatase (Ppx1p) prior to the labeling reaction. Ppx1p specifically degrades poly P and does not act on pyrophosphate or ATP (54). Figure 2D shows that we were able to label the poly(A) content of Δpho85 and Δpho80 RNA preparations following Ppx1p treatment and the observed distribution of poly(A) in the mutant strains appeared normal. No poly(A) was detected when reaction buffer replaced Ppx1p or when the Ppx1p was heat-denatured prior to use. Interestingly, the labeling efficiency of wild-type RNA following Ppx1p pre-treatment was also enhanced, demonstrating the presence of poly P also in wild-type RNA, consistent with our poly P measurements in wild-type strains (Figure 2B). Taken together the results from this figure show that poly P accumulated in Δpho85 and Δpho80 strains, that this metabolite co-purified with total RNA extracted from mutant and wild-type strains and that poly P was responsible for the observed inhibition of poly(A) polymerase.
Poly P binds poly(A) polymerase
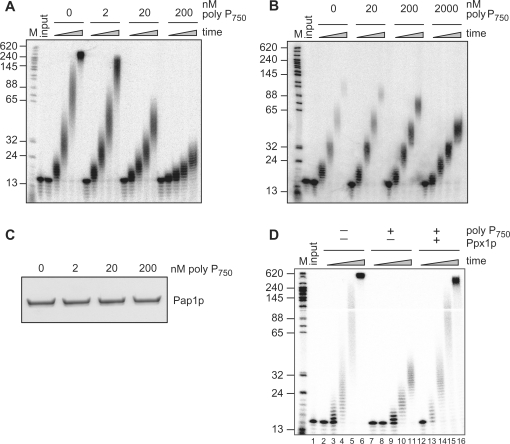
Since the poly(A)-labeling assay uses CoTP, poly(A) polymerase activity is restricted to a single round of nucleoside addition. To examine the effects of poly P under multiple turn-over conditions, we performed poly(A) polymerase assays with an end-labeled A15 RNA primer, cold ATP and recombinant yeast Pap1p (Figure 3A). Under the conditions employed, yeast Pap1p extended the primer to ∼300 to 400 adenosines in 60 min. In the presence of 2 nM poly P750, we observed a slight reduction of the length of the polyadenylation products during the time course. This effect was stronger with 20 nM poly P750 and 200 nM resulted in almost complete inhibition. Similar effects were observed when poly P of varying length (average of 28 and 61 phosphate residues) was tested (data not shown). The poly P concentrations used for these assays are likely to be physiologically relevant, as yeast nuclei are thought to harbor poly P concentrations of up to 89 μM (1).
Figure 3.
Poly P inhibition of poly(A) polymerase activity in vitro. (A) 32P-labeled A15 RNA primer was incubated with poly(A) polymerase and cold ATP in the absence or presence of the indicated amounts of poly P750. After 0, 5, 15, 30 and 60 min incubation time, an aliquot of the reaction was removed and analyzed on a 15% denaturing polyacrylamide gel. (A) Reaction with S. cerevisiae Pap1p and (B) reaction with bovine poly(A) polymerase. (C) Effects of poly P binding on the solubility of Pap1p. Following incubation of Pap1p with the indicated amounts of poly P750, reactions were cleared by centrifugation and soluble material was resolved by denaturing SDS–PAGE and stained with Coomassie Brilliant Blue. (D) Polyadenylation assay as described in (A). However, Pap1p was pre-incubated with H2O (lanes 2–6) or 200 nM poly P750 (lanes 7–16) for 15 min at 30°C. Subsequently, reaction mixtures were treated with H2O (lanes 2–11) or Ppx1p (lanes 12–16) for 30 min at 30°C and polyadenylation reactions were then started by the addition of A15 primer. HpaI-digested pBR322 fragments were 5′-end labeled and served as marker bands (M) with the indicated size.
Next, we tested whether poly P also inhibited the activity of bovine poly(A) polymerase. Under the employed conditions bovine Pap extended the A15 RNA primer by approximately 100 adenosines within 1 hour (Figure 3B). When 20 nM to 2 μM of poly P750 were included in the reaction, the length of the products was reduced but inhibition was not complete. Therefore, poly P was a less potent inhibitor of bovine poly(A) polymerase compared to the yeast enzyme.
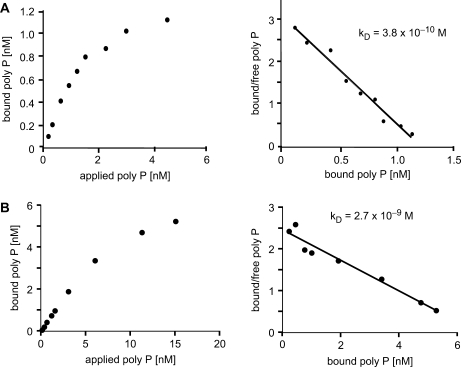
Poly P is an efficient chelator of divalent metal ions (1). A possible explanation for the inhibitory effect on poly(A) polymerase activity could be the chelation of magnesium ions which are essential for Pap1p function. However, the magnesium in our assays is present in large molar excess over inhibitory poly P concentrations (calculated as the amount of free inorganic phosphate). Therefore, we considered the possibility that poly P may inhibit the enzymes by direct binding. To test this, we performed filter-binding assays with radioactively labeled poly P750 and yeast or bovine poly(A) polymerase. We determined an apparent dissociation constant of 0.38 nM for the yeast enzyme and of 2.7 nM for the bovine enzyme (Figure 4). It has to be pointed out, however, that one poly P molecule is likely to provide multiple binding sites for Pap1p. Therefore, it seems possible that binding was influenced by avidity effects. Irrespective of that, the tight binding of poly P to poly(A) polymerase was most likely responsible for the observed inhibitory effects of poly P in polyadenylation assays.
Figure 4.
Binding of poly P to yeast and bovine poly(A) polymerase. Poly P binding by yeast (A) and bovine (B) poly(A) polymerase determined by filter-binding assay as a function of poly P750 concentration (left panels). Scatchard plot representations of the obtained data (right panels) were used to derive the indicated apparent kD values. The slope of the lines obtained equal −1/kD.
To address the mechanisms of inhibition, we considered the possibility that poly P binding may result in unspecific aggregation or precipitation of the enzyme. To test this idea we incubated Pap1p with increasing amounts of poly P, collected protein aggregates by centrifugation and resolved remaining soluble material by SDS–PAGE. As shown in Figure 3C, poly P binding did not result in precipitation of the enzyme. Thus, unspecific aggregation can be excluded as a possible reason for the observed inhibition. To test whether the inhibition of poly P on Pap1p activity was reversible, we pre-incubated Pap1p with 200 nM poly P to allow binding of poly P and Pap1p. As expected, this resulted in strong inhibition of polyadenylation activity (Figure 3D, lanes 7–11). However, Pap1p activity could be almost completely restored by treatment of the Pap1p–poly P complexes with Ppx1p (Figure 3D, lanes 12–16). This demonstrated that the inhibition of Pap1p by poly P was reversible.
The Pap1p that we used in the poly(A) length analysis and in the polyadenylation assay was purified from E. coli, which also contains poly P (1). Thus, purified Pap1p may already be partially bound to poly P causing a partial inhibition of the enzymatic activity. To test this, we performed a polyadenylation assays with Pap1p that was pretreated with Ppx1p. Since the treatment did not noticeably increase the activity of Pap1p preparation (data not shown), we conclude that most of Pap1p that we used in our experiments was not pre-bound by poly P. This indicated that the conditions used for over-expression of Pap1p did not induce significant poly P accumulation in E. coli.
Poly P accumulation contributes to cordycepin sensitivity
We showed that Δpho85 and Δpho80 strains are cordycepin hypersensitive, that these strains accumulated poly P and that poly P acted as an efficient inhibitor of poly(A) polymerase. These observations suggested a correlation between cellular poly P accumulation and the drug-dependent growth defect. Ogawa and co-workers (6) reported accumulation of poly P in strains mutant for PHM5, PHM6, PHM2 and CTF19 during the so-called ‘overplus’ conditions. However, we could not observe cordycepin-hypersensitive growth associated with these mutant strains (Figure 5A). Moreover, our poly P measurements in these strains did not indicate a significant increase of the polymer under conditions, which revealed a strong increase in Δpho85 and Δpho80 strains (Figure 5B). The differences between the measurements presented here and in Ogawa et al. are possibly due to differences in growth conditions at the time of measurement. Furthermore, our recent genome-wide assessment of poly P metabolism failed to identify other mutants that would result in poly P accumulation comparable to levels observed in Δpho85 and Δpho80 strains (38). It seems possible, therefore, that cordycepin-hypersensitive growth is a consequence of elevated poly P levels exceeding a certain threshold in Δpho85 and Δpho80 strains and that such levels are not achieved or stably maintained in other mutants defective in poly P metabolism.
To further test whether cordycepin sensitivity was due to poly P accumulation in Δpho85 and Δpho80 mutants, we interfered with the ability of the strains to synthesize poly P and asked whether this alleviated cordycepin sensitivity. When low concentrations of phosphate (0.1 mM) are present in the medium growth of the yeast is only marginally reduced, but no significant poly P accumulation can be measured (32). We tested Δpho80 strains on plates containing cordycepin but low phosphate and found that Δpho80 strains showed significant resistance to cordycepin under these conditions (Figure 5C). This observation suggested that abundant phosphate was required for the observed cordycepin hypersensitivity. Furthermore, the PHO pathway regulates poly P accumulation in yeast (6). Figure 5D shows that cellular levels of poly P were significantly reduced in Δpho2, Δpho4 and Δpho84 mutants compared to wild type. We produced double mutants of Δpho80 and other components of the PHO pathway and found that Δpho80 Δpho4, Δpho80 Δpho2 and Δpho80 Δpho84 double mutants did not accumulate poly P (Figure 5D). The cellular poly P content of the analyzed double mutants correlated well with a loss of cordycepin hypersensitivity associated with the single Δpho80 mutation (Figure 5C). These observations are consistent with the proposal that poly P accumulation in Δpho80 strains contributed to cordycepin hypersensitivity. Increased cellular poly P levels may interfere with poly(A) polymerase activity, and this may cause hypersensitive growth in the presence of cordycepin, another inhibitor of RNA synthesis.
To further test this idea we asked whether a temperature-sensitive mutation in the 3′ end formation factor Rna14p (rna14-1) can duplicate the effects observed with cordycepin in Δpho80 mutants. It was previously shown that combination of mutations in rna14 and pap1 resulted in a synthetic growth defect (39) and rna14 mutant strains are cordycepin hypersensitive [(46); our unpublished data]. Figure 5E shows that combination of the Δpho80 mutation with the rna14-1 allele indeed resulted in synthetic lethality. This genetic interaction strongly supported the proposal that accumulation of poly P in Δpho80 strains affected 3′ end formation via inhibition of poly(A) polymerase activity and that this inhibition in combination with another defect in 3′ end formation led to the observed synthetic growth defect.
DISCUSSION
We provide evidence for an unexpected link between poly P metabolism and polyadenylation in yeast. We propose that a synergistic effect of poly P-mediated inhibition of poly(A) polymerase together with inhibition of RNA synthesis by CoTP can cause lethality. Synthetic lethality observed between PHO80 and an essential factor involved in 3′ end formation underscores this assumption. Poly P may therefore play a role in the regulation of poly(A) polymerase activity.
The starting point of this work was an unbiased genome-wide screen for mutants that showed hypersensitive and/or resistant growth in the presence of cordycepin. The rational for this screen was that cordycepin will be converted to CoTP, which will then provoke RNA-chain termination in the nucleus. Although CoTP will terminate RNA synthesis when incorporated during transcription, it seems possible that polyadenylation would be a more sensitive target since up to 70 consecutive adenosines are incorporated during this process. This may be particularly pronounced under the low cordycepin conditions (40 μg/ml) that were employed during our screening; as expected, mutants in 3′ end formation factors displayed hypersensitive growth in the presence of such cordycepin concentrations (Figure 1A and data not shown). Although it remains unclear which other cellular ATP-consuming activities are sensitive to cordycepin or CoTP, cordycepin-hypersensitive growth can be indicative of a functional involvement of a mutation in 3′ end formation and poly(A) metabolism. A complete account of the results obtained from our cordycepin-dependent growth screening will be presented elsewhere (S.H. and B.D., unpublished data). However, no additional links emerged between cordycepin hypersensitivity and poly P metabolism other than the one presented here for Δpho85 and Δpho80 strains.
The kinase activity of the Pho85p/Pho80p CDK–cyclin complex controls phosphate-dependent gene expression by regulating both the activity and localization of the Pho4p transcription factor (50,55). Interestingly, poly P was proposed to act as a phosphate buffer that can be mobilized to maintain constant internal phosphate levels under conditions where external phosphate levels fluctuate (7,8) and the PHO pathway was linked to expression of genes that are involved in poly P homeostasis (6). As a consequence, constitutive activation of the PHO pathway (e.g. in Δpho80 strains) was expected to result in increased poly P levels. However, Ogawa and co-workers (6) could not observe increased levels of poly P in Δpho80 strains. In contrast, it was reported that Δpho85 strains have increased poly P (52). We re-evaluated the poly P content of Δpho85 and Δpho80 strains and detected an approximately 3-fold increase in both strains compared to wild-type (Figure 2B). We attribute these conflicting poly P measurements to fluctuations of poly P levels in the yeast depending on the growth state and recently proposed a standardized procedure for poly P determination (32).
We found that the presence of increased amounts of co-purified poly P in RNA preparations from Δpho85 and Δpho80 strains was accompanied by a strong inhibition of poly(A) polymerase activity. Consistent with our observations, previous biochemical work on the effects of poly P on yeast Pap1p revealed an inhibitory activity on tRNA primed poly(A) synthesis (56). We show in this work by filter-binding assays that poly P directly binds to yeast Pap1p with a kD in the low nanomolar range (0.38 nM). This value is comparable to the kD of 0.48 nM that was determined for the poly P–Lon interaction, which stimulates Lon protease activity (16). Pap1p and Lon are thus the interactors with the highest reported affinities for poly P. Consistent with a physiological relevant role for the interaction of poly P and Pap1p, nuclear poly P levels were reported to reach micromolar concentrations (1); furthermore, nuclei predominantly contain poly P of an average chain length of approximately 45 phosphate residues (33), and poly P with similar length (poly P28 and poly P61) acted as efficient inhibitor of Pap1p in our in vitro experiments (Figure 2C). We have not delineated the binding site of poly P on Pap1p, but potential interaction surfaces include regions of RNA primer binding or of ATP binding (57,58). Notably, binding of poly P to bovine poly(A) polymerase and inhibition of its activity is approximately 10-fold weaker compared to the yeast enzyme. This may have important consequences on a potential in vivo interaction of poly P and poly(A) polymerases in higher eukaryotes.
What is the biological role of poly P-mediated inhibition of poly(A) polymerase activity? We predict that such a function must be connected to growth conditions that are accompanied by high poly P concentrations and that the control of poly P levels is at least partially under control of the PHO pathway. We recently showed that poly P content reaches a peak when cells experience the diauxic shift following depletion of glucose from the medium (32). This growth phase is characterized by major changes in the gene expression program (59,60). It seems possible that the down-regulation of many hundreds of genes during diauxic shift is accompanied by an inhibition of polyadenylation. In this context, it is worth pointing out that cellular poly(A) levels inversely correlate with poly P levels when cells enter diauxic shift (32,60). Our observation that the inhibitory effects of poly P on Pap1p were completely reversible (Figure 3D) may indicate that poly P action can be transient and controlled through a dynamic nuclear poly P metabolism (33). Furthermore, fluctuations of nuclear poly P levels may interfere with RNA processing in a microenvironment as was suggested for the regulation of the bacterial degradosome (26). Since no global polyadenylation defect could be observed at steady state in Δpho80 and Δpho85 strains (Figure 2D) poly P may act on specific gene loci and not as a general suppressor of Pap1p activity. A deeper understanding of poly P biochemistry and metabolism will be required to address these possibilities in future experiments.
ACKNOWLEDGEMENTS
We want to thank Georges Martin (Biozentrum, Basel, Switzerland) for the generous gift of purified yeast and bovine poly(A) polymerases and C. Jakob and N. Amrhein for important discussions and support. This work was funded by Swiss National Science Foundation (3100A0-112083/1 to F.M.F.) and (PP00A-102941 to B.D.). Funding to pay the Open Access publication charges for this article was provided by Swiss National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kornberg A. Inorganic polyphosphate: a molecule of many functions. Prog. Mol. Subcell. Biol. 1999;23:1–18. doi: 10.1007/978-3-642-58444-2_1. [DOI] [PubMed] [Google Scholar]
- 2.Kulaev IS, Vagabov VM, Kulakovskaya TV. The Biochemistry of Inorganic Polyphosphates. 2nd. Ltd, Chichester, West Sussex: John Wiley and Sons; 2004. [Google Scholar]
- 3.Bonting CF, Kortstee GJ, Zehnder AJ. Properties of polyphosphate: AMP phosphotransferase of Acinetobacter strain 210A. J. Bacteriol. 1991;173:6484–6488. doi: 10.1128/jb.173.20.6484-6488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsieh PC, Shenoy BC, Jentoft JE, Phillips NF. Purification of polyphosphate and ATP glucose phosphotransferase from Mycobacterium tuberculosis H37Ra: evidence that poly(P) and ATP glucokinase activities are catalyzed by the same enzyme. Protein Expr. Purif. 1993;4:76–84. doi: 10.1006/prep.1993.1012. [DOI] [PubMed] [Google Scholar]
- 5.Martinez P, Zvyagilskaya R, Allard P, Persson BL. Physiological regulation of the derepressible phosphate transporter in Saccharomyces cerevisiae. J. Bacteriol. 1998;180:2253–2256. doi: 10.1128/jb.180.8.2253-2256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogawa N, DeRisi J, Brown PO. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell. 2000;11:4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neef DW, Kladde MP. Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol. Cell. Biol. 2003;23:3788–3797. doi: 10.1128/MCB.23.11.3788-3797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas MR, O'Shea EK. An intracellular phosphate buffer filters transient fluctuations in extracellular phosphate levels. Proc. Natl Acad. Sci. USA. 2005;102:9565–9570. doi: 10.1073/pnas.0501122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood HG, Clark JE. Biological aspects of inorganic polyphosphates. Annu. Rev. Biochem. 1988;57:235–260. doi: 10.1146/annurev.bi.57.070188.001315. [DOI] [PubMed] [Google Scholar]
- 10.Bol DK, Yasbin RE. Characterization of an inducible oxidative stress system in Bacillus subtilis. J. Bacteriol. 1990;172:3503–3506. doi: 10.1128/jb.172.6.3503-3506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pick U, Weiss M. Polyphosphate hydrolysis within acidic vacuoles in response to amine-induced alkaline stress in the halotolerant alga Dunaliella salina. Plant Physiol. 1991;97:1234–1240. doi: 10.1104/pp.97.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao NN, Kornberg A. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 1996;178:1394–1400. doi: 10.1128/jb.178.5.1394-1400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ault-Riche D, Fraley CD, Tzeng CM, Kornberg A. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 1998;180:1841–1847. doi: 10.1128/jb.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao NN, Liu S, Kornberg A. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J. Bacteriol. 1998;180:2186–2193. doi: 10.1128/jb.180.8.2186-2193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 16.Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science. 2001;293:705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- 17.Kumble KD, Kornberg A. Inorganic polyphosphate in mammalian cells and tissues. J. Biol. Chem. 1995;270:5818–5822. doi: 10.1074/jbc.270.11.5818. [DOI] [PubMed] [Google Scholar]
- 18.Jacinto E, Hall MN. Tor signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell. Biol. 2003;4:117–126. doi: 10.1038/nrm1018. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Fraley CD, Faridi J, Kornberg A, Roth RA. Inorganic polyphosphate stimulates mammalian TOR, a kinase involved in the proliferation of mammary cancer cells. Proc. Natl Acad. Sci. USA. 2003;100:11249–11254. doi: 10.1073/pnas.1534805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiba T, Nishimura D, Kawazoe Y, Onodera Y, Tsutsumi K, Nakamura R, Ohshiro M. Modulation of mitogenic activity of fibroblast growth factors by inorganic polyphosphate. J. Biol. Chem. 2003;278:26788–26792. doi: 10.1074/jbc.M303468200. [DOI] [PubMed] [Google Scholar]
- 21.Kawazoe Y, Shiba T, Nakamura R, Mizuno A, Tsutsumi K, Uematsu T, Yamaoka M, Shindoh M, Kohgo T. Induction of calcification in MC3T3-E1 cells by inorganic polyphosphate. J. Dent. Res. 2004;83:613–618. doi: 10.1177/154405910408300806. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez-Ruiz L, Gonzalez-Garcia I, Castro C, Brieva JA, Ruiz FA. Inorganic polyphosphate and specific induction of apoptosis in human plasma cells. Haematologica. 2006;91:1180–1186. [PubMed] [Google Scholar]
- 23.Han KY, Hong BS, Yoon YJ, Yoon CM, Kim YK, Kwon YG, Gho YS. Polyphosphate blocks tumour metastasis via anti-angiogenic activity. Biochem. J. 2007;406:49–55. doi: 10.1042/BJ20061542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusano S, Ishihama A. Functional interaction of Escherichia coli RNA polymerase with inorganic polyphosphate. Genes Cells. 1997;2:433–441. doi: 10.1046/j.1365-2443.1997.13203301320330.x. [DOI] [PubMed] [Google Scholar]
- 25.Shiba T, Tsutsumi K, Yano H, Ihara Y, Kameda A, Tanaka K, Takahashi H, Munekata M, Rao NN, et al. Inorganic polyphosphate and the induction of rpoS expression. Proc. Natl Acad. Sci. USA. 1997;94:11210–11215. doi: 10.1073/pnas.94.21.11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blum E, Py B, Carpousis AJ, Higgins CF. Polyphosphate kinase is a component of the Escherichia coli RNA degradosome. Mol. Microbiol. 1997;26:387–398. doi: 10.1046/j.1365-2958.1997.5901947.x. [DOI] [PubMed] [Google Scholar]
- 27.McInerney P, Mizutani T, Shiba T. Inorganic polyphosphate interacts with ribosomes and promotes translation fidelity in vitro and in vivo. Mol. Microbiol. 2006;60:438–447. doi: 10.1111/j.1365-2958.2006.05103.x. [DOI] [PubMed] [Google Scholar]
- 28.Indge KJ. Polyphosphates of the yeast cell vacuole. J. Gen. Microbiol. 1968;51:447–455. doi: 10.1099/00221287-51-3-447. [DOI] [PubMed] [Google Scholar]
- 29.Urech K, Durr M, Boller T, Wiemken A, Schwencke J. Localization of polyphosphate in vacuoles of Saccharomyces cerevisiae. Arch. Microbiol. 1978;116:275–278. doi: 10.1007/BF00417851. [DOI] [PubMed] [Google Scholar]
- 30.Lichko L, Kulakovskaya T, Pestov N, Kulaev I. Inorganic polyphosphates and exopolyphosphatases in cell compartments of the yeast Saccharomyces cerevisiae under inactivation of PPX1 and PPN1 genes. Biosci. Rep. 2006;26:45–54. doi: 10.1007/s10540-006-9003-2. [DOI] [PubMed] [Google Scholar]
- 31.Kulaev IS, Vagabov VM, Kulakovskaya TV, Lichko LP, Andreeva NA, Trilisenko LV. The development of A. N. Belozersky's ideas in polyphosphate biochemistry. Biochemistry. 2000;65:271–278. [PubMed] [Google Scholar]
- 32.Werner TP, Amrhein N, Freimoser FM. Novel method for the quantification of inorganic polyphosphate (iPoP) in Saccharomyces cerevisiae shows dependence of iPoP content on the growth phase. Arch. Microbiol. 2005;184:129–136. doi: 10.1007/s00203-005-0031-2. [DOI] [PubMed] [Google Scholar]
- 33.Lichko LP, Kulakovskaya TV, Kulaev IS. Inorganic polyphosphate and exopolyphosphatase in the nuclei of Saccharomyces cerevisiae: dependence on the growth phase and inactivation of the PPX1 and PPN1 genes. Yeast. 2006;23:735–740. doi: 10.1002/yea.1391. [DOI] [PubMed] [Google Scholar]
- 34.Lichko LP, Andreeva NA, Kulakovskaya TV, Kulaev IS. Exopolyphosphatases of the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2003;3:233–238. doi: 10.1016/S1567-1356(02)00205-2. [DOI] [PubMed] [Google Scholar]
- 35.Lichko LP, Kulakovskaia TV, Kulaev IS. Characteristics of Saccharomyces cerevisiae nuclear polyphosphatase activity. Biokhimiia. 1996;61:488–495. [PubMed] [Google Scholar]
- 36.Sethuraman A, Rao NN, Kornberg A. The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2001;98:8542–8547. doi: 10.1073/pnas.151269398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wurst H, Shiba T, Kornberg A. The gene for a major exopolyphosphatase of Saccharomyces cerevisiae. J. Bacteriol. 1995;177:898–906. doi: 10.1128/jb.177.4.898-906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freimoser FM, Hurlimann HC, Jakob CA, Werner TP, Amrhein N. Systematic screening of polyphosphate (poly P) levels in yeast mutant cells reveals strong interdependence with primary metabolism. Genome Biol. 2006;7:R109. doi: 10.1186/gb-2006-7-11-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minvielle-Sebastia L, Preker PJ, Keller W. Rna14 and Rna15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- 40.Martin G, Keller W. Tailing and 3′ end labeling of RNA with yeast poly(A) polymerase and various nucleotides. RNA. 1998;4:226–230. [PMC free article] [PubMed] [Google Scholar]
- 41.Wahle E. Purification and characterization of a mammalian polyadenylate polymerase involved in the 3′ end processing of messenger RNA precursors. J. Biol. Chem. 1991;266:3131–3139. [PubMed] [Google Scholar]
- 42.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akiyama M, Crooke E, Kornberg A. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. J. Biol. Chem. 1992;267:22556–22561. [PubMed] [Google Scholar]
- 44.Maale G, Stein G, Mans R. Effects of cordycepin and cordycepintriphosphate on polyadenylic and ribonucleic acid-synthesising enzymes from eukaryotes. Nature. 1975;255:80–82. doi: 10.1038/255080a0. [DOI] [PubMed] [Google Scholar]
- 45.Horowitz B, Goldfinger BA, Marmur J. Effect of cordycepin triphosphate on the nuclear DNA-dependent RNA polymerases and poly(A) polymerase from the yeast Saccharomyces cerevisiae. Arch. Biochem. Biophys. 1976;172:143–148. doi: 10.1016/0003-9861(76)90059-x. [DOI] [PubMed] [Google Scholar]
- 46.Minvielle-Sebastia L, Winsor B, Bonneaud N, Lacroute F. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate; sequence analysis reveals an RNA-binding domain in the Rna15 protein. Mol. Cell. Biol. 1991;11:3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwashima A, Kawasaki Y, Nosaka K, Nishimura H. Effect of thiamin on cordycepin sensitivity in Saccharomyces cerevisiae. FEBS Lett. 1992;311:60–62. doi: 10.1016/0014-5793(92)81367-u. [DOI] [PubMed] [Google Scholar]
- 48.Russnak R, Nehrke KW, Platt T. REF2 encodes an RNA-binding protein directly involved in yeast mRNA 3′ end formation. Mol. Cell. Biol. 1995;15:1689–1697. doi: 10.1128/mcb.15.3.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 50.Lenburg ME, O'Shea EK. Signaling phosphate starvation. Trends Biochem. Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- 51.Carroll AS, O'Shea EK. Pho85 and signaling environmental conditions. Trends Biochem. Sci. 2002;27:87–93. doi: 10.1016/s0968-0004(01)02040-0. [DOI] [PubMed] [Google Scholar]
- 52.McDonald AE, Niere JO, Plaxton WC. Phosphite disrupts the acclimation of Saccharomyces cerevisiae to phosphate starvation. Can. J. Microbiol. 2001;47:969–978. doi: 10.1139/w01-099. [DOI] [PubMed] [Google Scholar]
- 53.Auesukaree C, Homma T, Tochio H, Shirakawa M, Kaneko Y, Harashima S. Intracellular phosphate serves as a signal for the regulation of the PHO pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:17289–17294. doi: 10.1074/jbc.M312202200. [DOI] [PubMed] [Google Scholar]
- 54.Wurst H, Kornberg A. A soluble exopolyphosphatase of Saccharomyces cerevisiae. Purification and characterization. J. Biol. Chem. 1994;269:10996–11001. [PubMed] [Google Scholar]
- 55.Komeili A, O'Shea EK. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 56.Sillero MA, de Diego A, Silles E, Osorio H, Sillero A. Polyphosphates strongly inhibit the tRNA dependent synthesis of poly(A) catalyzed by poly(A) polymerase from Saccharomyces cerevisiae. FEBS Lett. 2003;550:41–45. doi: 10.1016/s0014-5793(03)00815-9. [DOI] [PubMed] [Google Scholar]
- 57.Martin G, Keller W, Doublie S. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 2000;19:4193–4203. doi: 10.1093/emboj/19.16.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balbo PB, Bohm A. Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis. Structure. 2007;15:1117–1131. doi: 10.1016/j.str.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 60.Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]