Abstract
The widely accepted model for the processing of tRNAs in Escherichia coli involves essential initial cleavages by RNase E within polycistronic transcripts to generate pre-tRNAs that subsequently become substrates for RNase P. However, recently we identified two polycistronic tRNA transcripts whose endonucleolytic processing was solely dependent on RNase P. Here we show that the processing of the secG leuU and metT leuW glnU glnW metU glnV glnX polycistronic transcripts takes place through a different type of maturation pathway. Specifically, RNase P separates the tRNA units within each operon following the endonucleolytic removal of the distal Rho-independent transcription terminator, primarily by RNase E. Failure to remove the Rho-independent transcription terminator inhibits RNase P processing of both transcripts leading to a decrease in mature tRNA levels and dramatically increased levels of full-length transcripts in an RNase E deletion strain. Furthermore, we show for the first time that RNase G also removes the Rho-independent transcription terminator associated with the secG leuU operon. Our data also demonstrate that the Rne-1 protein retains significant activity on tRNA substrates at the non-permissive temperature. Taken together it is clear that there are multiple pathways involved in the maturation of tRNAs in E. coli.
INTRODUCTION
The processing of tRNA genes in Escherichia coli has been studied extensively over the past 15 years (1–9). Based on these experiments, a general model for the maturation of tRNAs, particularly those contained within polycistronic operons, has emerged in which the primary transcripts are presumed to be cleaved by RNase E to yield pre-tRNAs that are subsequently matured by RNase P at their 5′ ends and by a combination of RNase T, RNase PH, RNase BN, RNase II and RNase D at their 3′ ends (5,10).
However, the observed differential efficiency of RNase E cleavages among a large number of tRNA precursors as well as the absence of consensus RNase E cleavage sites in some of them (6,7,10) raised the possibility that alternative processing pathways existed. In fact, the recent analysis of the valV valW and leuQ leuP leuV operons has demonstrated a new processing pathway in which RNase P is the only endonuclease involved in the separation of tRNAs within these polycistronic transcripts (10).
In the experiments described here, we sought to determine if additional endoribonucleases, other than RNase E and RNase P, are involved in tRNA maturation. These include RNase G (encoded by rng), which is homologous to the catalytic domain of RNase E (11) and has been shown to be involved in the maturation of the 5′ end of the 16S rRNA (12,13) and to serve as a less efficient alternative to RNase E for initiating the decay of some mRNAs in wild-type cells (14). RNase Z (encoded by rnz) is known to be essential for the maturation of tRNA precursors that do not contain a chromosomally encoded CCA determinant in eukaryotes (15,16), archaea (17) and certain prokaryotes (18). However, since all of the tRNA genes in E. coli contain chromosomally encoded CCA sequences (19), it seemed likely that its RNase Z ortholog functioned in a different pathway. In fact, it has now been implicated in the decay of mRNAs, suggesting that it also functions as an alternative enzyme for RNase E in certain aspects of RNA metabolism (20). Similarly, RNase LS (encoded by rnlA), which is involved in the degradation of T4 late-gene mRNAs, has been suggested to play a role in E. coli RNA metabolism (21).
We now show that the processing of secG leuU and metT (metT leuW glnU glnW metU glnV glnX) polycistronic transcripts is accomplished through a distinct tRNA processing pathway involving multiple endonucleases. In this pathway, RNase E and/or RNase G-dependent removal of the Rho-independent transcription terminator significantly improves the efficiency of RNase P-mediated processing of the polycistronic transcripts. However, unlike with leuT and leuZ (5), the RNase E cleavages are not absolutely essential for processing to take place, in part because of the ability of RNase G to remove the transcription terminator and/or RNase P to process the full-length transcripts, including the terminator, inefficiently.
MATERIALS AND METHODS
Bacterial strains and plasmids
The E. coli strains used in this study were all derived from MG1693 (thyA715 rph-1), which was provided by the E. coli Genetic Stock Center (Yale University), and are listed in Table 1. The rne-1 and rnpA49 alleles encode temperature-sensitive RNase E and RNase P proteins, respectively, that are unable to support cell viability at 44°C (22–24). SK3564 [rneΔ1018::bla thyA715 rph-1 recA56 srlD::Tn10/pDHK30(rng-219 Smr/Spr)/pWSK129 (Kmr)] is an RNase E deletion strain in which cell viability is supported by a mutant RNase G (rng-219) protein synthesized from a single copy plasmid. The details of the construction of this strain will be published separately (D. Chung, Z. Min, B.-C. Wang and S.R. Kushner, manuscript in preparation).
Table 1.
Strains used in this study
| Strains | Genotype | Reference/source |
|---|---|---|
| MG1693 | thyA715 rph-1 | B. Bachmann |
| SK2525 | rnpA49 thyA715 rph-1 rbsD296::Tn10 Tcr | (7) |
| SK2534 | rne-1 rnpA49 thyA715 rph-1 rbsD296::Tn10 Tcr | (7) |
| SK2538 | rng::cat thyA715 rph-1 Cmr | (14) |
| SK2541 | rne-1 rng::cat thyA715 rph-1 Cmr | (14) |
| SK3166 | rne-1 ΔrnlA::kan thyA715 rph-1 Kmr | Perwez and Kushner, unpublished data |
| SK3170 | ΔrnlA::kan thyA715 rph-1 Kmr | Perwez and Kushner, unpublished data |
| SK3564 | rneΔ1018::bal thyA715 rph-1 recA56 srlD::Tn10/pDHK30 (rng219 Smr/Spr)/pWSK129 (Kmr) | Chung et al., manuscript in preparation |
| SK5665 | rne-1 thyA715 rph-1 | (24) |
| SK9795 | Δrnz::kan thyA715 rph-1 Kmr | (20) |
| SK9797 | rne-1 Δrnz::kan thyA715 rph-1 Kmr | (20) |
Growth of bacterial strains and isolation of total RNA
The growth of the bacterial strains used in this work as well as the isolation, quantification and normalization of RNA samples have been described previously (10). Northern analyses were carried out as outlined by Mohanty and Kushner (10).
Primer extension experiments
Primer extension analysis of the secG leuU transcript was carried out essentially as described previously (10) with the following modifications. The nucleotide sequences were obtained from a PCR DNA product (amplified from wild-type genomic DNA using primers upstream and downstream of the secG leuU operon) using the primer SECG-227 (primer a, Figure 1A), which was also used for the reverse transcription reaction.
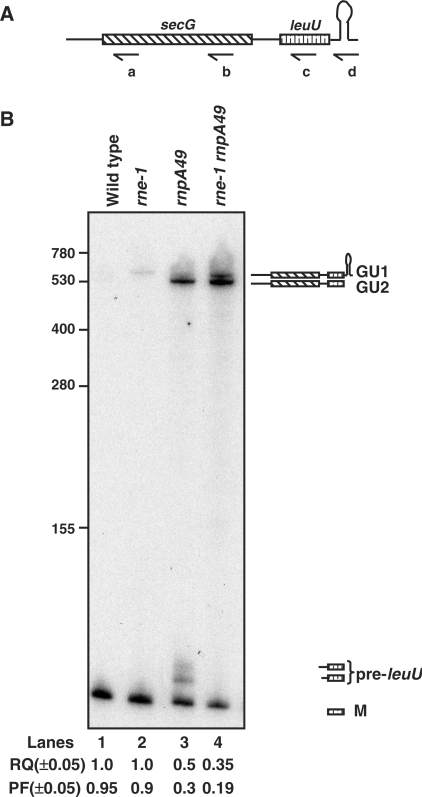
Figure 1.
Analysis of the processing of secG leuU transcript. (A) Schematic representation of secG leuU operon (not drawn to scale). Relative positions of the oligonucleotide probes (a: SECG-227, b: SECG3′, c: LEUU and d: LEUU-TER) used in the northern analysis are shown below the diagram. (B) Northern analysis of secG leuU transcript. Total RNA (12 μg/lane) was separated on 6% PAGE, transferred to a nylon membrane and probed multiple times as described in the Materials and Methods. Only the blot probed with c is shown. The genotypes of the strains used are noted above each lane. The deduced structures and the names for the processing intermediates of secG leuU transcript are shown to the right. The RQ of the mature tRNALeu2 (M) in the various genetic backgrounds was calculated by setting the wild-type level at 1.0. PF denotes the fraction of the mature tRNA relative to the total amount of both processed and unprocessed species in each specific genetic background. RQ and PF were obtained from the average of at least three independent experiments. The RNA size standards (nucleotide) (Invitrogen) are shown to the left.
RT-PCR cloning and sequencing of 5′–3′ ligated transcripts
The 5′ and 3′ ends of various transcripts described in the text were identified by cloning and sequencing the RT-PCR products obtained from 5′ to 3′ end-ligated circular RNAs (25–28). Total steady-state RNA (3 μg) was self-ligated in the presence of T4 RNA ligase (NEB). For analysis of the secG leuU and secG transcripts, total RNA was initially treated with tobacco acid pyrophosphatase (TAP) according to the manufacturer's instructions (Epicentre Technologies, Madison, WI, USA) to convert the 5′-triphosphate termini to phosphomonoesters prior to self-ligation. The circular transcripts were reverse transcribed using a gene-specific primer close to the 5′–3′ junction in the presence of Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's specifications. The 5′–3′ junctions of the resulting cDNAs were amplified using either a pair of gene-specific primers (secG, leuU, hisR and cysT) or one upstream and one downstream gene-specific primers (secG leuU) in the presence of JumpStart REDTaq™ DNA polymerase (Sigma). All the primers used in the cDNA amplifications were engineered to contain a suitable restriction site for directed cloning into pWSK29 (29). DNA sequencing was carried out using an automated sequencer (Applied Biosystems 3730×l DNA analyzer).
Oligonucleotide probes and primers
The sequences of the primers used in the experiments reported here are available on request.
RESULTS
Processing of the secG leuU transcript is RNase P dependent
The secG leuU operon is unique in E. coli in that it contains both a protein encoding gene (secG) and a tRNA gene (leuU) (Figure 1A). The product of the leuU gene (tRNALeu2) recognizes the CUC and CUU leucine codons and is essential for cell viability (30). The secG leuU transcript is terminated with a Rho-independent transcription terminator and the two genes are separated by a 14 nt spacer region that has been predicted to contain RNase P cleavage site based on in vitro experiments (31). Initially, we examined the processing of this transcript in wild-type, rne-1, rnpA49 and rne-1 rnpA49 strains by northern analysis using a series of oligonucleotides (Figure 1A-a, b, c and d). For the sake of simplicity, only the results with the leuU-specific probe (Figure 1A-c) are shown (Figure 1B).
An expected band of 87 nt, corresponding to the mature tRNALeu2 (M) was observed in all the strains (Figure 1B, lanes 1 and 4), with its level primarily a function of RNase P activity. Thus the loss of RNase P activity led to reductions of ∼50% in the relative quantity (RQ) and ∼70% in the processed fraction (PF) of mature tRNA compared to the wild-type strain (Figure 1B, lanes 1 and 3). The reduction in the level of mature tRNALeu2 in the rnpA49 mutant was consistent with the accumulation of significant amounts of a high-molecular weight processing intermediate (GU2) and a mixture of pre-tRNALeu2 species (pre-leuU) that were 6–11 nt longer than the mature tRNALeu2 (Figure 1B, lane 3).
In contrast, inactivation of RNase E had only a minor effect on either the RQ or PF of tRNALeu2 compared to the wild-type strain (Figure 1B, lanes 1 and 2). However, a different high-molecular weight intermediate (GU1) that was ∼40 nt longer than GU2 appeared in the rne-1 strain, but was only present in very low levels (Figure 1B, lane 2). When both RNase E and RNase P were inactivated there was a further reduction of the RQ and PF of the mature tRNA to ∼0.35 and ∼0.19, respectively, compared to the wild-type control along with a concomitant increase in both the GU1 and GU2 species (Figure 1B, lane 4). In addition, the pre-leuU species that were present in the rnpA49 single mutant (Figure 1B, lane 3) disappeared, suggesting that they were generated by RNase E cleavages in the intergenic region between secG and leuU. In the wild-type control, both the GU1 and GU2 species were only visible after a much longer exposure of the membrane (data not shown).
A secG-specific probe (Figure 1A-a) hybridized only to bands GU1 and GU2 (data not shown) indicating that these transcripts contained both secG and leuU-coding sequences. Surprisingly, a band corresponding to the full-length secG mRNA (∼420 nt) was only visible after a much longer exposure (data not shown).
The two larger secG leuU transcripts (GU1 and GU2) have identical 5′ termini
Although the data presented in Figure 1 clearly indicated that the two larger transcripts contained both secG and leuU-coding sequences, it was not clear what accounted for their difference in electrophoretic mobility. We hypothesized that the smaller transcript (GU2) was derived from larger GU1 species by endonucleolytic cleavage(s) either at the 5′ or 3′ end. Accordingly, we analyzed the 5′ ends of the secG leuU transcripts employing primer extension analysis (Figure 2A). A strong primer extension product (I) terminating at 81 nt (G) and a second weaker primer extension product (II) (∼10% of I) terminating at 84 nt (C) upstream of the putative secG translation start codon were detected in the wild-type strain. Since no other longer primer extension products were observed, I and II represented the major and minor transcription initiation sites, respectively, for the secG leuU transcript. The sequence upstream of these transcription start sites (I and II) contained consensus (4/6) −10 and −35 sequences of a σ70 promoter (Figure 2B).
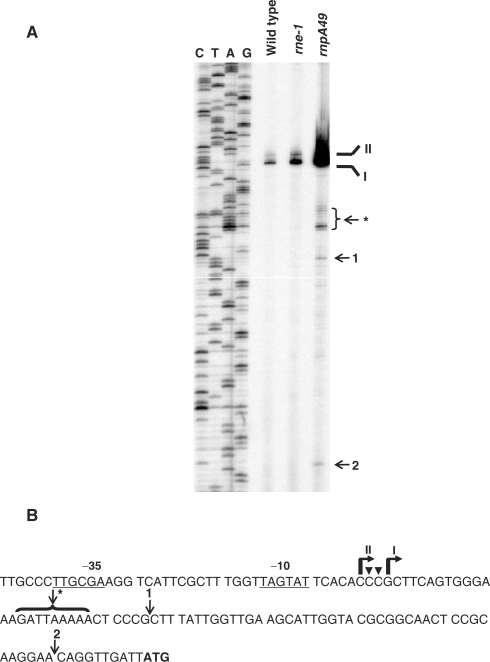
Figure 2.
Primer extension analysis of the promoter region of secG leuU transcript. (A) Autoradiograph of the primer extension analysis. The reverse transcription products of total RNA (10 μg/lane) isolated from wild-type (MG1693), rne-1 (SK5665) and rnpA49 (SK2525) strains using the primer a (Figure 1A) complementary to the sequences 54 nt downstream of translation start codon (ATG, B) were separated on a 6% PAGE as described in the Materials and Methods section. The CTAG sequencing reactions were carried out using the same primer and a secG leuU PCR DNA fragment as template. The two 5′ ends (I and II) of secG leuU transcript are indicated. Arrows indicate 5′ termini derived from the presumed endonucleolytic cleavages. Asterisk (*) indicates the 5′ ends identified using RT-PCR cloning and sequencing analysis (Figure 4). (B) The nucleotide sequences of the upstream of secG leuU translation start (bold) site showing the transcription start sites (bent arrows) and endonucleolytic processing (arrows) sites. The G (I) at −81 and C (II) at −84 nt upstream of the translation initiation site are the major transcription initiation sites. However, based on the RT-PCR cloning and sequencing of self-ligated transcripts (Figure 4), some transcription initiation also takes place at −83 and −82 nt (downward arrowheads). The putative −10 and −35 sequences (underlined) are shown.
More importantly, the major primer extension products (I and II) in both the rne-1 and rnpA49 mutants were at positions identical to those observed in the wild-type strain (Figure 2A), suggesting that transcription initiation started at the same locations in all genetic backgrounds. Consistent with the northern analysis (Figure 1B), the amount of primer extension products increased significantly in both the rne-1 and rnpA49 mutants compared to the wild-type strain. The additional minor primer extension products visible in the rnpA49 strain (arrows and asterisk in Figure 2A) presumably arise from endonucleolytic cleavages downstream of the two transcription initiation sites.
The full-length secG leuU transcript that includes the Rho-independent transcription terminator only accumulates in the absence of RNase E
Since both larger processing intermediates (GU1 and GU2) still retained the 5′ transcription start site (Figure 2), we surmised that the difference in their electrophoretic mobility arose from a processing event at the 3′ end. As shown in Figure 1A, the secG leuU operon contains a strong Rho-independent transcription terminator predicted to be 36 nt in length. Since the nucleotides immediately downstream of the leuU CCA determinant were A/U-rich, we suspected that RNase E might be responsible for removal of the terminator region. To test this hypothesis directly, we probed northern blots with an oligonucleotide (Figure 1A-d) specific to the terminator region of the secG leuU transcript. As predicted, only the larger of the two species (GU1) was detected in the strains tested, indicating that it represented the full-length secG leuU transcript (data not shown). While the full-length transcript was very weakly visible in the wild-type and rnpA49 strains after long exposure, there was an ∼16-fold increase in the rne-1 strain and ∼32-fold in the rne-1 rnpA49 double mutant compared to the wild-type control (Table 2), indicating a significant role of RNase E in the generation of the secG leuU species that lacked the terminator (GU2), which accumulated in the rnpA49 mutant (Figure 1A, lane 3).
Table 2.
RQ of secG leuU full-length transcripts and PF of mature tRNALeu2 in various strains
| Strain | Genotype | RQa | PFb |
|---|---|---|---|
| MG1693 | Wild type | 1 | 0.95 ± 0.05 |
| SK5665 | rne-1 | 16 ± 2 | 0.95 ± 0.05 |
| SK3564 | rneΔ1018/rng-219 | 98 ± 10 | 0.5 ± 0.05 |
| SK2525 | rnpA49 | 1.5 ± 0.2 | 0.3 ± 0.05 |
| SK2534 | rne-1 rnpA49 | 32 ± 2 | 0.3 ± 0.05 |
| SK2538 | rng::cat | 1 ± 0.1 | 0.95 ± 0.05 |
| SK2541 | rne-1 rng::cat | 29 ± 2 | 0.7 ± 0.05 |
| SK9795 | Δrnz::kan | 1 ± 0.1 | 0.95 ± 0.05 |
| SK9797 | rne-1 Δrnz::kan | 20 ± 2 | 0.9 ± 0.05 |
| SK3170 | ΔrnlA::kan | 1 ± 0.1 | 0.95 ± 0.05 |
| SK3166 | rne-1 ΔrnlA::kan | 12 ± 2 | 0.95 ± 0.05 |
However, the importance of the RNase E cleavage for removal of the terminator from the secG leuU transcript was not completely clear from the above experiment because the PF of mature tRNALeu2 did not change significantly in the rne-1 strain compared to the wild-type control (Figure 1B, lanes 1 and 2, Table 2). If the removal of the terminator by RNase E were a prerequisite for RNase P processing of the secG leuU transcript, we expected to see comparable reductions in the PF of the mature tRNALeu2 in both the rne-1 and rnpA49 strains (Figure 1B, lanes 2 and 3, Table 2).
We, therefore, hypothesized that either there was residual RNase E activity in the rne-1 mutant at the non-permissive temperature or that additional endoribonucleases were involved in the removal of the terminator, particularly because there was also ∼10-fold difference between the level of the full-length transcript (GU1) in the rne-1 strain compared to the terminator-less species (GU2) in the rnpA49 mutant (Figure 1B). To address the possibility of residual RNase E activity in the rne-1 strain, we tested an rne deletion strain [rneΔ1018, (32)] in which cell viability was restored by a mutant RNase G protein (encoded by rng-219, D. Chung, Z. Min, B.-C. Wang and S. R. Kushner, manuscript in preparation). In this case, the RQ of GU1 increased ∼98-fold compared to the wild-type strain (Table 2). More importantly, the PF of the mature tRNALeu2 decreased to ∼50% of the wild-type level (Table 2). In addition, a small amount of a processing intermediate containing the leuU tRNA with the terminator still attached was now present in this genetic background (data not shown), indicating that either RNase P or the mutant RNase G protein could inefficiently process the full-length transcript at or upstream of the 5′ end of leuU.
To investigate the potential involvement of other endoribonucleases, we measured the level of the full-length transcript (GU1) in a series of mutants defective in RNase E, RNase G (14), RNase LS (21) and RNase Z (20). While the level of the full-length secG leuU transcripts (GU1) in the rng, rnlA and rnz single mutants was identical to the wild-type strain within experimental error (Table 2), it increased significantly in the rne-1 rng double mutant compared to the rne-1 single mutant (Table 2).
The stability of the secG leuU transcript is dependent on RNase P processing
The significant difference in the steady-state levels of the secG leuU transcript with terminator (GU1) in the rne-1 strain and without terminator (GU2) in the rnpA49 mutant (Figure 1B, lanes 2 and 3) suggested that processing of the secG leuU transcript was highly dependent on an RNase P cleavage event. Accordingly, we determined the half-lives of both secG leuU transcripts by using either leuU- or secG-specific probes (Figure 1A-a and c). Since both probes yielded identical results, only the results obtained with probe c (leuU-specific) are shown in Figure 3.
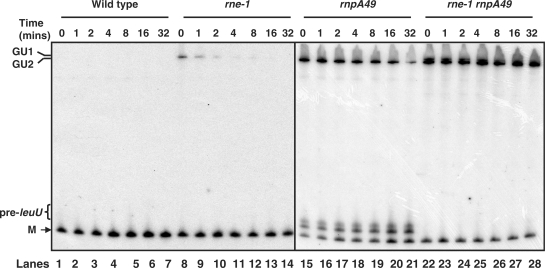
Figure 3.
Decay of the secG leuU transcript. Total RNA (12 μg/lane) from various strains was isolated at times (minutes after rifampicin addition) indicated at the top of the blot and was separated on a 6% PAGE, transferred to a nylon membrane and probed with probe c (Figure 1A) as described in the Materials and Methods section. Two autoradiograms (one for wild-type and rne-1 and another for rnpA49 and rnpA49 rne-1 strains) are shown side-by-side. GU1, GU2, pre-leuU and M are as described in Figure 1.
As expected, neither of the secG leuU transcripts was detected in the wild-type strain (Figure 3, lanes 1–7), indicating a half-life of <30 s (10). The full-length secG leuU transcript (GU1) had a half-life of 1.5 ± 0.4 min in the rne-1 strain (Figure 3, lanes 8–14). In contrast, in the rnpA49 mutant the terminator-less secG leuU species (GU2) was significantly stabilized with a half-life of 17 ± 2 min (Figure 3, lanes 15–21). When both the RNase E and RNase P were inactivated (rne-1 rnpA49), the half-lives of both the GU1 and GU2 transcripts increased further to 3 ± 0.5 and >32 min, respectively (Figure 3, lanes 22–28). Furthermore, in the rne-1 rnpA49 double mutant, the amount of the GU2 species increased slowly during the course of the experiment (Figure 3, lanes 22–28). Also of interest was that the level of pre-tRNA (pre-leuU) species in the rnpA49 mutant increased ∼2-fold during the course of the experiment (Figure 3, lanes 15–21), while their complete absence in the rne-1 rnpA49 double mutant (lanes 22–28) was consistent with them being derived from inefficient RNase E cleavages upstream of leuU.
Separation of the secG mRNA from leuU by RNase P leads to its rapid degradation
While the data described in Figures 1 and 3 clearly showed that processing of the secG leuU transcript was primarily dependent on the action of RNase P, surprisingly no significant amount of secG transcript was detected using either secG-specific oligonucleotides (probes a and b, Figure 1A) or a full-length secG DNA probe in all the strains tested (wild-type, rne-1, rneΔ1018/rng-219, rnpA49 and rne-1 rnpA49, rne-1 rng::cat, rne-1 rnz::kan and rne-1 rnlA::kan, data not shown). These observations indicated that the secG mRNA was rapidly degraded with a half-life of <30 s, after cleavage of the secG leuU transcript by RNase P.
RNase E cleaves in the A/U-rich region downstream of leuU CCA mature terminus
Although the primer extension and northern blot analysis revealed that the full-length secG leuU transcript (GU1) was processed in part by RNase E to GU2, we were interested in determining the actual cleavage site(s). We employed an RT-PCR technique using 5′–3′ end-ligated transcripts (25) to clone and sequence both the 5′ and 3′ termini of the transcripts simultaneously. Since both the secG leuU transcripts appeared to retain the 5′ end of the primary transcript (Figure 2), the triphosphate was converted to monophosphate employing TAP treatment to facilitate the self-ligation of 5′ and 3′ ends of transcripts in presence of T4 RNA ligase.
Consistent with the primer extension experiments (Figure 2), the majority of the 5′ ends of the transcripts were located at either 81 nt (11/15 in the wild-type strain, 8/14 in the rne-1 and 7/16 in the rnpA49 mutant) or in a limited number of cases at 82–84 nt upstream of the putative translation start codon (Figure 4A–D). Three of the clones (1/14 in the rne-1 and 2/16 in the rnpA49 mutants) had 5′ ends at 66 and 59 nt upstream of the translation start (Figure 4C and D), which corresponded to primer extension products seen in Figure 2 (marked with asterisk). These particular termini were also identified in clones generated from self-ligated transcripts independent of prior TAP treatment, suggesting that these ends most likely arose from endonucleolytic cleavages (data not shown). In contrast, none of the 5′ ends at −81 to −84 nt were detected in self-ligated transcripts in the absence of treatment with TAP (data not shown), indicating that the majority of these ends had retained a 5′ triphosphate. In fact, the presence of only a very limited number of primary transcripts with 5′-phosphomonoesters were indicated, since there was a >100-fold reduction in the level of PCR amplification products from the self-ligated transcripts in the absence of TAP treatment (data not shown).
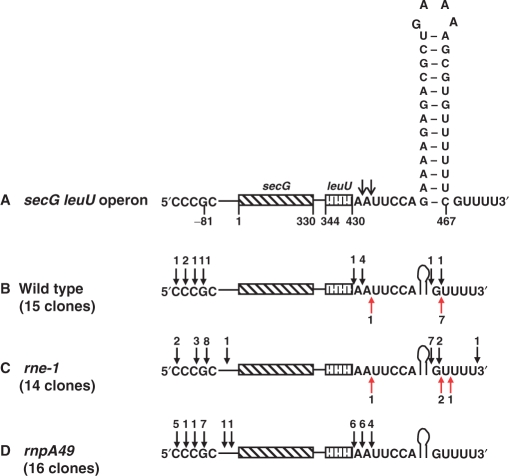
Figure 4.
Identification of 5′ and 3′ ends of secG leuU transcripts using RT-PCR cloning of 5′–3′ self-ligated transcripts. Total steady-state RNA was self-ligated, reverse transcribed, amplified using secG and leuU-specific primers, cloned and sequenced as described in the Materials and Methods section. (A) Schematic representation of secG leuU transcript showing the 5′ and 3′ nucleotide sequences. A predicted Rho-independent transcription terminator at the 3′ end is shown. The nucleotide at the beginning of the translation start codon (AUG) is numbered as 1. The open arrows represent the major endonucleolytic cleavages downstream of leuU CCA terminus based on this study (also see Figure 5C). The 5′ and 3′ ends of secG leuU transcripts are identified in wild type (B) rne-1 (C) and rnpA49 (D) strains. Each closed arrow (both black and red) above nucleotides at the 5′ end and after a nucleotide at the 3′ end represents a 5′ and 3′ end, respectively, of the secG leuU transcript. A red closed arrow indicates the presence of 1–5 non-templated A's at the positions marked. The numbers directly above or below each arrow represent the number of clones identified with these respective 5′ or 3′ ends.
Sixty percent (9/15) of the 3′ ends of the secG leuU transcripts in the wild-type strain occurred after the Rho-independent transcription terminator (Figure 4B). However, transcripts from 6/15 (40%) wild-type clones ended at or after 1–2 residues downstream of the mature CCA terminus within the AAUU sequence (Figure 4B, 430–432 nt). When RNase E was inactivated, the number of the 3′ ends of the secG leuU transcripts terminating after the Rho-independent transcription terminators increased to 93% (13/14) (Figure 4C). The lone transcript in the rne-1 mutant that had a terminus 2 nt downstream from the CCA (Figure 4C) may have arisen from a cleavage by RNase G, as suggested from the data presented in Table 2.
Furthermore, the secG leuU transcripts from all of the 16 clones derived from the rnpA49 mutant were missing the terminator and ended at 430—432 nt (Figure 4D), indicating that the RNase E and/or RNase G cleavages occurred within the AAUU sequence (Figure 4A). Finally, it should be noted that the number of secG leuU transcripts, with and without a transcription terminator, obtained in this experiment reflect the distribution of particular species in each genetic background, in agreement with the northern analysis shown in Figure 1. Interestingly, ∼24% of all the clones contained post-transcriptionally added A residues of up to 5 nt (Figure 4B and C, red arrows).
A single RNase P cleavage of the secG leuU transcript releases tRNALeu2 with a mature 5′ terminus
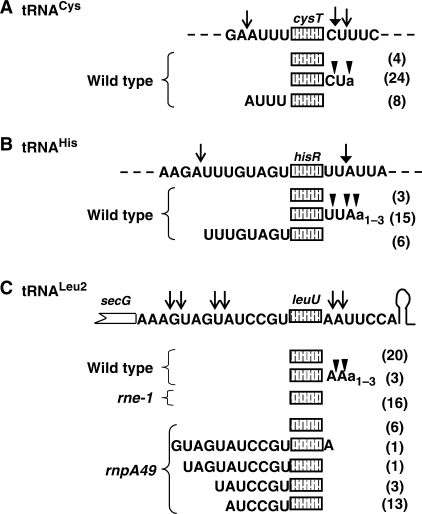
The data described in Figures 1 and 3 suggested that RNase P, not RNase E, was essential for the removal of tRNALeu2 from the secG leuU polycistronic transcript. To provide further support for this conclusion, we mapped the 5′ and 3′ ends of tRNALeu2 in various genetic backgrounds by cloning and sequencing of 5′–3′ end-ligated junctions. As controls, we also identified the 5′ and 3′ ends of tRNAHis (hisR) and tRNACys (cysT) using identical techniques, since both of these tRNAs are present in single copy, are part of polycistronic transcripts, and have been shown to be dependent on RNase E for their initial processing (5,7).
In the case of tRNALeu2, we expected that all of the clones would have mature 5′ ends in a wild-type strain because the RNase P cleavage would generate them directly. In contrast, with tRNAHis and tRNACys, we hypothesized that some of them would have 5′ extensions resulting from initial upstream cleavages by RNase E. As predicted, all the tRNALeu2 clones (23/23) in the wild-type strain had the mature 5′ end (Figure 5C). Three of these clones (3/23, 13%) had immature 3′ ends occurring 1–2 nt downstream of the CCA terminus in the AAUU sequence (Figure 5C). Similarly, 3′ termini were also obtained with the secG leuU-specific transcripts (Figure 4, 431–432 nt).
Figure 5.
Identification of 5′ and 3′ ends of tRNACys (A), tRNAHis (B) and tRNALeu2 (C) using RT-PCR cloning of 5′–3′ end-ligated transcripts. The total RNA was self-ligated, reverse transcribed, amplified using cysT, hisR and leuU-specific primers, cloned and sequenced as described in the Materials and Methods section. A schematic representation of each tRNA with up and downstream sequences is shown directly above respective tRNA clones. RNase E cleavage sites as identified previously (solid arrow) and in this study (open arrow) are indicated at the top of each diagram. Multiple 3′ ends (inverted triangles) with similar 5′ ends were obtained for various tRNA clones. Numbers in the parenthesis to the right of each diagram are the number of clones sequenced for that species. Lower case a's indicate the presence of non-templated residues.
In contrast, 6/24 (25%) of the tRNAHis and 8/36 (22%) of the tRNACys clones had immature 5′ ends in the wild-type strain (Figure 5A and B). These immature termini were 8 and 4 nt upstream of the tRNAHis and tRNACys mature 5′ termini, respectively, in what appeared to be RNase E consensus cleavage sites (A/U-rich regions). Furthermore, ∼63% (15/24) of the tRNAHis and ∼67% (24/36) of the tRNACys clones had immature 3′ ends that were up to 2 nt (U or UU for tRNAHis and C or CU for tRNACys) downstream of the mature CCA terminus. The sites of the 5′ and 3′ immature termini of tRNAHis and tRNACys were consistent with the previous reports of RNase E-processing sites to release these pre-tRNAs from their respective polycistronic transcripts (5). The presence of a higher percentage of unprocessed 3′ ends of tRNAHis (∼63%) and tRNACys (∼67%) compared to tRNALeu2 (∼13%) in the wild-type strain suggests the possibility of differential rates of 3′ end maturation by RNase T, RNase D and RNase BN in this genetic background (MG1693 contains the rph-1 allele, which inactivates RNase PH). Interestingly, ∼9% of the tRNALeu2 clones and ∼20–25% of both the tRNAHis and tRNACys clones contained 1–3 nt of untemplated A residues downstream of the immature terminus (Figure 5A–C).
Since we observed some pre-leuU species in the rnpA49 single mutant, but not in the rne-1 rnpA49 double mutant (Figure 1B, lanes 3 and 4), we also mapped the 5′ and 3′ ends of tRNALeu2 in an rnpA49 single mutant. Under these circumstances, 18/24 (75%) of the tRNALeu2 clones contained immature 5′ ends with 6–11 unprocessed nucleotides (Figure 5C). There were four different termini within a 6-nt region (GUAGUA) of the 14 nt secG leuU intercistronic spacer (Figure 5C), indicative of inefficient, non-specific cleavages by RNase E upstream of tRNALeu2 under conditions in which the secG leuU transcript accumulated in the absence of RNase P. Furthermore, almost all of the tRNALeu2 clones (23/24) had mature 3′ ends even if their 5′ ends were not processed by RNase P in the rnpA49 mutant (Figure 5C).
To obtain further evidence that RNase P cleavage of the secG leuU transcript directly generated the mature 5′ terminus of the tRNALeu2, we examined the 3′ termini of secG transcripts. We hypothesized that a single RNase P cleavage of secG leuU transcript to release the mature 5′ end of tRNALeu2 would yield secG transcripts containing the intact 14 nt spacer present between secG and leuU (Figure 5C). In fact, when we mapped the 3′ termini of secG mRNAs, ∼20% (8/40) of the clones contained the entire 14 nt spacer in both the wild-type and rne-1 strains (data not shown). The rest of the clones contained secG transcripts with 3′ ends distributed throughout the coding sequence (data not shown).
Processing of the metT operon
We subsequently turned our attention to leuW (tRNALeu3), which is part of the metT operon (metT leuW glnU glnW metU glnV glnX) (Figure 6A). In addition to the presence of a leucine tRNA, this operon was of interest because Li et al. (33) observed recently that the steady-state level of all the methionine (met) tRNA transcripts increased significantly in an rnpA49 mutant. Accordingly, we analyzed the processing of this complex operon, employing the nine distinct oligonucleotide probes (a–i) shown in Figure 6A.
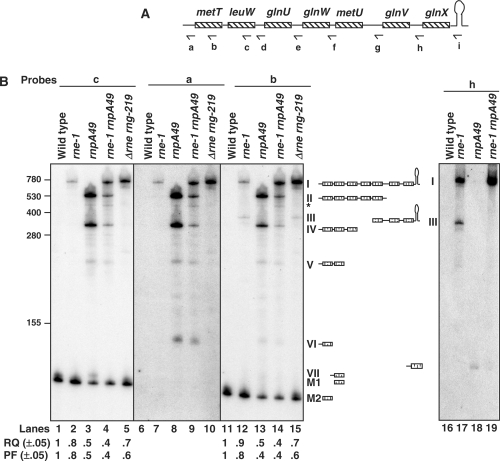
Figure 6.
Analysis of processing of the metT polycistronic transcript. (A) Schematic presentation of metT operon (not drawn to scale). Relative positions of the oligonucleotide probes (a: METT-UP, b: METT-265, c: LEUW-338, d: GLNU-UP, e: GLNW-UP, f: METU-UP, g: GLNV-UP, h: GLNX-UP and i: GLNX-TER) used in the northern analysis are shown below the cartoon. (B) Northern analysis of metT leuW glnU glnW metU glnV glnX transcripts. Total RNA (12 μg/lane) was separated on 6% PAGE, transferred to a nylon membrane and probed multiple times with probes a–i (A) as described in Materials and Methods section. The genotypes of the strains used are noted above each lane. The deduced structures and the names for the processing intermediates are shown. The weak band marked with a asterisk (*) is due to either a non-specific or weak cleavage by an unidentified ribonuclease in the middle of the metU-coding sequence, since it hybridized to the probes a–f (A and B, lanes 1–15, data not shown) but not to the probes g–i (B, lanes 16–19, data not shown). RQ and PF of the mature tRNAMet (M2) and tRNALeu2 (M1) were calculated as described in Figure 1. The RNA size standards (nucleotide) (Invitrogen) are shown to the left. Mature tRNALeu3 (M1) is 85 nt in length while the mature tRNAMet (M2) is 79 nt.
As the leuW-coding sequence is unique to this operon and differs significantly from the other seven leucine tRNAs, we initially probed the northern blot with the probe c (Figure 6A). As expected a band corresponding to the mature tRNALeu3 (M1) was observed in all the strains (Figure 6B, lanes 1–5). While both the RQ and PF of the mature tRNALeu3 were reduced only marginally in the rne-1 mutant compared to the wild-type control, the decreases were much more significant in both the rnpA49 and rne-1 rnpA49 mutants. Besides the mature tRNA band (M1), five other processing intermediates (I, II, IV, V and VII, Figure 6B, lanes 2–4) were observed in various genetic backgrounds.
In order to determine the composition of these intermediates, we hybridized the blot with two additional oligonucleotides (Figure 6A-a and b) that were complementary to the 5′ non-coding and tRNAMet (metT and metU)-coding regions, respectively. Both of these probes hybridized to bands I, II, IV and V but not to band VII (Figure 6B, lanes 8–9 and 13–14), which was thus identified as leuW with an unprocessed 5′ end that occurred in the absence of RNase P (Figure 6B, lane 3). However, both the probes a and b hybridized to a new processing intermediate (VI) of ∼125 nt that was present in rnpA49 mutants (lanes 8–9 and 13–14) and was absent in the c-probed blot. Thus species VI represented metT containing an unprocessed 5′ end.
Furthermore, probe b hybridized to an additional high-molecular weight processing intermediate (III) in the rne-1 mutant (lane 12), and, as expected, also hybridized to the mature tRNAMet in all genetic backgrounds (M2, Figure 6B, lanes 11–15). Similar to what was observed with tRNALeu3 (M1), inactivation of RNase E led to only a small reduction in the relative level of the mature tRNAMet (lane 12). In contrast, the loss of RNase P activity had a much greater effect on the amount of mature tRNAMet (Figure 6B, lanes 13 and 14).
Probing the blot with oligonucleotide i (Figure 6A), which was complementary to the terminator sequences of the operon, identified bands I and III (Figure 6B) in all strains defective in RNase E (data not shown). Since these bands were missing in the rnpA49 single mutant (Figure 6B, data not shown), RNase E appeared to be primarily responsible for removal of the terminator. However, the level of band I increased significantly in the rne-1 rnpA49 double mutant (Figure 6B, lanes 2, 4, 7, 9, 12 and 14), suggesting roles for both RNase E and RNase P in initiating the processing of this transcript. As was seen with the secG leuU transcript (Table 2), the level of the full-length transcript that retained the downstream Rho-independent transcription terminator sequences of the metT operon increased ∼5-fold along with a concomitant reduction in the PF of mature tRNAMet and tRNALeu3 in an rne deletion mutant compared to the temperature-sensitive rne-1 mutant (Figure 6B, lanes 2, 5, 7, 10, 12 and 15, data not shown).
The composition of all the processing intermediates shown in Figure 6B was further confirmed using the additional oligonucleotide probes d, e, f, g and h (Figure 6A). For example, oligonucleotides g and h (Figure 6A), which were specific for the intergenic regions between metU, glnV and glnX, hybridized to bands I and III in the rne-1 single mutant but only to band I in rne-1 rnpA49 double mutant (Figure 6B, lanes 17 and 19, data not shown). In the rnpA49 mutant, the only intermediate observed was glnX containing extra nucleotides at its 5′ terminus with probe h (Figure 6B, lane 18). Similarly, while probe d hybridized to bands I, II and IV, probe e hybridized to bands I and II only (data not shown). Furthermore, probe f (Figure 6A) did not hybridize to band III containing metU (Figure 6B, data not shown) suggesting that it was missing the 5′ upstream sequences.
Taken together, we conclude that the band I (Figure 6B) represents the full-length transcript containing all seven tRNAs. Bands II, IV, V and VI are processing intermediates that retain the 5′ non-coding region along with one or more tRNA species. Thus band II has the first five tRNAs (metT leuW glnU glnW metU), band IV has the first three tRNAs (metT leuW glnU), band V had two tRNAs (metT leuW) and band VI has only metT tRNA. On the other hand, band III is derived from the 3′ end of the transcript by an RNase P cleavage at 5′ end of metU and contains metU glnV glnX as well as the intact Rho-independent transcription terminator.
DISCUSSION
Previous studies have identified two distinct tRNA processing pathways in E. coli involving either RNase E or RNase P as the primary endoribonuclease responsible for generating pre-tRNAs from polycistronic transcripts (5,10). In the RNase E-dependent pathway, polycistronic tRNA transcripts are separated into pre-tRNAs solely by RNase E with the pre-tRNAs being subsequently matured at their 5′ termini by RNase P (5). In contrast, in the RNase P-dependent pathway, polycistronic tRNA transcripts are separated into pre-tRNAs by RNase P, generating the mature 5′ ends in a single step independent of RNase E (10). In this report, we describe for the first time an alternative tRNA processing pathway for the secG leuU and metT (metT leuW glnU glnW metU glnV glnX) polycistronic transcripts (Figure 7A and B) that involves certain aspects of both the RNase E- and RNase P-dependent pathways. What makes this pathway distinct in wild-type E. coli is that the removal of the Rho-independent transcription terminator associated with each polycistronic transcript, primarily by RNase E, significantly stimulates their rapid processing by RNase P into pre-tRNAs containing mature 5′ termini. However, RNase E is not absolutely required for this step, in part because RNase G can also remove the terminator sequences and RNase P can inefficiently process a full-length transcript that retains the terminator. The role of RNase G in the removal of the terminator is partially masked by the fact that there is ∼10 times more RNase E in the cell than RNase G (34).
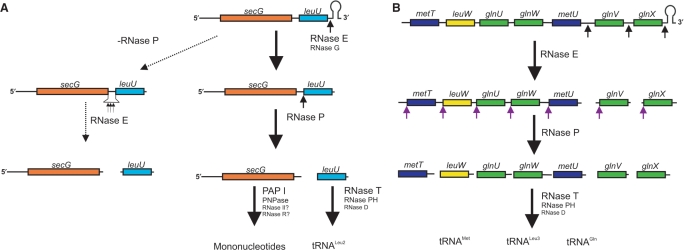
Figure 7.
Processing pathways for the secG leuU and metT polycistronic transcripts in E. coli. (A) Processing of the secG leuU transcript. The Rho-independent transcription terminator is removed, primarily by RNase E, but can also be cleaved by RNase G. Subsequently, RNase P separates the leuU sequence from secG generating the mature 5′ end of tRNALeu2 and leaving 14 nt downstream of the translation stop codon of secG. Final maturation of the 3′ terminus of leuU is carried out by a combination of RNase T, RNase PH and RNase D (38) to generate a mature tRNALeu2. The secG mRNA is rapidly degraded exonucleolytically in a process that is dependent primarily on polynucleotide phosphorylase (PNPase) and poly(A) polymerase (PAP I). However, we cannot rule out a role for either RNase II or RNase R at this time. In the absence of RNase P, RNase E can inefficiently cleave at multiple locations with the 14 nt spacer region between secG and leuU. However, as noted in Figure 1, only a very small fraction of the full-length secG leuU transcript is susceptible to this reaction. (B) Processing of the metT polycistronic transcript. RNase cleavages remove the Rho-independent transcription terminator and separates glnV and glnX from the rest of the transcript. RNase P is required to separate the five upstream tRNAs. Subsequent 3′ end processing by RNase T, RNase PH and RNase D generate mature tRNALeu3, tRNAMet and tRNAGln.
Evidence for RNase P being the primary ribonuclease involved in the separation of tRNALeu2 from secG (Figure 7A) was derived from the fact that secG leuU transcripts lacking the Rho-independent transcription terminator (GU2) accumulated in the rnpA49 single mutant but not in either the rne-1 (Figure 1B) or rneΔ1018 deletion strains (data not shown). Furthermore, the identification of secG mRNA fragments with the intact 14 nt secG leuU intergenic sequence along with no unprocessed 5′ nucleotides in the pre-tRNALeu2 species (Figure 5, data not shown) confirmed that a single-RNase P cleavage generated the mature 5′ terminus of tRNALeu2 and separated it from the upstream secG mRNA.
Similarly, inactivation of RNase P led to the dramatic accumulation of several large precursor species of the metT operon (bands II and IV) that contained either the first five (metT leuV glnU glnW metU) or first three (metT leuW glnU) tRNAs, respectively, (Figure 6), suggesting that these tRNAs were also endonucleolytically processed primarily by RNase P (Figure 7B). The significant drop in the RQ and PF of the mature tRNALeu2, tRNALeu3 and tRNAMet species in rnpA49 mutants (Figures 1B and 6B, data not shown) was consistent with this model (Figure 7).
Interestingly, the presence of a Rho-independent transcription terminator appears to significantly inhibit the ability of RNase P to process both transcripts. This was evidenced by the accumulation of full-length transcripts with intact terminators only in RNase E-deficient strains as opposed to either no accumulation in the wild-type strain or only accumulation of transcripts with the terminator removed in the RNase P-deficient strain (Figures 1 and 6, Table 2). Thus, the role of RNase E and/or RNase G for the processing of the secG leuU and metT polycistronic transcripts is primarily to remove the Rho-independent transcription terminator, thereby facilitating the RNase P-dependent cleavages that generate pre-tRNAs with mature 5′ termini. This is in marked contrast to the requirement for initial RNase E cleavages to generate pre-tRNAs with immature 5′ ends in the processing of the glyW and argX polycistronic transcripts (5,7).
Although it is not clear at this time why the presence of the Rho-independent transcription terminator inhibits RNase P activity, it appears from the data presented in Table 2 that under normal physiological conditions, the terminator sequences are endonucleolytically removed by RNase E and/or RNase G prior to RNase P action to generate the pre-tRNAs. In addition, the last three tRNAs in the complex metT operon are also separated by RNase E as suggested by the accumulation of band III (metU glnV glnX) in the rne-1 strain (Figure 6B). This species was generated by an RNase P cleavage at the mature 5′ terminus of metU (Figure 7B, data not shown). The disappearance of this band in the rnpA49 single mutant and rnpA49 rne-1 double mutants supports this interpretation.
An unexpected observation was the involvement of RNase G in the removal of the Rho-independent transcription terminator associated with the secG leuU transcript (Figure 4, Table 2). This is the first demonstration that RNase G plays any role in the processing of E. coli transcripts containing tRNAs. While the removal of the Rho-independent transcription terminator associated with the metT polycistronic transcript appeared independent of RNase G activity (data not shown), it seems likely that other polycistronic tRNA transcripts terminated in a Rho-independent fashion may also be partially processed by RNase G.
Even though the presence of the Rho-independent transcription terminator inhibited processing by RNase P, the relatively low steady-state level of full-length transcripts of both operons in the rne-1 strain indicates that RNase P can process the full-length transcript, although more slowly than in the presence of functional RNase E. This was consistent with a significant increase in the half-life of full-length secG leuU transcript (Figure 3) and the level of both the secG leuU and metT operon full-length transcripts (Figures 1 and 6) in the rne-1 rnpA49 double mutant compared to either of the single mutants. These results indicate that either RNase E or RNase P can independently initiate the processing of these transcripts, although removal of the Rho-independent transcription terminator is normally the preferred first step in a wild-type strain.
Our data also clearly show that the temperature-sensitive Rne-1 protein, which is the most widely used allele to characterize which substrates are degraded in an RNase E-dependent fashion, clearly retains significant activity on tRNA substrates at the non-permissive temperature. Thus, there was a 5–6-fold increase in the transcript level in the rneΔ1018 strain compared to the rne-1 strain (2 h after shift to 44°C) for both the secG leuU and metT operons as well as concomitant reductions in the PF levels of tRNALeu2, tRNALeu3 and tRNAMet (Figure 6B, Table 2). An independent study from this laboratory has shown identical results for the argX (argX hisR leuT proM) and glW (glyW cysT leuZ) operons when comparing the rne-1 and rneΔ1018 strains (D. Chung, Z. Min, B.-C. Wang and S. R. Kushner, manuscript in preparation).
Previous microarray analysis of the E. coli transcriptome in an rnpA49 mutant led to the hypothesis that inactivation of RNase P specifically destabilized transcripts downstream of its cleavage sites (31). However, the dramatic destabilization of the secG mRNA after RNase P processing of the secG leuU transcript (Figures 1 and 3) clearly demonstrates that this idea is not entirely correct. In addition, the failure to stabilize the secG mRNA in any of the endonuclease mutants tested suggests that once the RNase P cleavage occurs, the upstream secG mRNA is rapidly degraded exonucleolytically.
Interestingly, a very low level of RNase E cleavages within the intergenic regions of both operons was observed in the absence of RNase P. Thus, the pre-leuU species were the result of RNase E cleavages upstream of leuU in the secG leuU transcript (Figures 1B, lane 3 and 7A). These cleavages were inefficient (Figure 1B, lane 3), occurred at multiple sites, and probably did not take place in wild-type cells (Figures 5C and 7A). Similarly for metT operon, the presence of precursors (bands II, IV, V and VI) in the rnpA49 single mutant and the reduction in their intensity in the rne-1 rnpA49 double mutant (Figure 6B) suggest that RNase E can also cleave in the intercistronic regions among the first five tRNAs with varying efficiencies. For example, the presence of a large amount of band IV indicates efficient cleavage between glnU and glnW, while the absence of an intermediate containing the first four tRNAs (metT leuW glnU glnW) suggests that RNase E is unable to cleave in the intercistronic region between glnW and metU. Similar inefficient cleavages of RNase P-dependent tRNA transcripts by RNase E have also been observed in the valV valW and leuQ leuP leuV operons (10).
With the work presented here, RNase E cleavage sites and relative efficiencies of cleavage are now available for 19/86 tRNA species (Table 3). This data is of particular interest because previous work has suggested the importance of an A/U-rich region in the cleavage site (35,36) and the presence of a G residue at the −2 position and either C or U at the +2 position (37). Of the 12 high-efficiency RNase E cleavage sites only one has a G at the −2 position. In contrast, 9/12 sites have a C or U at the +2 position (Table 3). In addition, it does not appear that RNase E can cleave closer than 1 nt downstream of the CCA determinant, even if the next nucleotide is a U.
Table 3.
Analysis of RNase E cleavage sites in tRNA precursors
| tRNA gene | 3′ terminal region | Efficiency of RNase E cleavage | Source/reference |
|---|---|---|---|
| lysY | CCAG↓UUUUA | High | (7) |
| tyrT | CCAU↓AAUUC | High | (5) |
| hisR | CCAUU↓AUUA | High | (5), This study |
| cysT | CCAC↓U↓UUCU | High | (5), This study |
| metU | CCAAAUUCU | High | This study |
| aspT | CCACCCUA↓A | High | Mohanty and Kushner, unpublished data |
| trpT | CCAGA↓AAUC | High | Mohanty and Kushner, unpublished data |
| proK | CCAAA↓CGAA | High | Mohanty and Kushner, unpublished data |
| lysW | CCAG↓UUUUA | High | Mohanty and Kushner, unpublished data |
| leuU | CCAA↓A↓UUCC | High | This study |
| glnV | CCAAUUUAU | High | This study |
| glnX | CCACAUUAA | High | This study |
| metT | CCAUCUUUU | Medium | This study |
| glnU | CCAUCUUCU | Medium | This study |
| leuQ | CCAAAAACC | Low | (10) |
| leuP | CCAAACGAG | Low | (10) |
| glnW | CCAUCGAAG | Very Low | This study |
| leuW | CCAUUCACC | Very Low | This study |
| valV | CCAUCCUGC | None | (10) |
Downward arrows indicate RNase E cleavage sites that have been mapped.
Based on the experiments reported above and our recent observations with the valV valW and leuQ leuP leuV operons (10), it is now clear that a significant number of tRNA precursors are dependent on RNase P for their initial separation from polycistronic transcripts rather than RNase E. In fact, a preliminary analysis indicates that the metZ operon (metZ metW metV) is also highly dependent on RNase P for processing (data not shown) as predicted from the microarray data of Li et al. (33). These observations provide further support for our hypothesis that the essential function of RNase P may be related to its requirement for the processing of polycistronic tRNA transcripts rather than the processing of the 5′ ends of pre-tRNAs (10). Finally, it would not be surprising if there were, in fact still yet, unidentified additional pathways for the maturation of specific tRNAs. Thus the mechanisms of tRNA maturation in E. coli are far more diverse than has previously been envisioned.
ACKNOWLEDGEMENTS
We are indebted to V. Maples, T. Perwez and D. Chung for invaluable technical assistance in the construction of various multiple mutants used in this work. This work was supported in part by a grant from the National Institutes of General Medical Sciences (GM57220) to S.R.K. Funding to pay the Open Access publication charges for this article was provided by GM57220.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kelly KO, Deutscher MP. The presence of only one of five exoribonucleases is sufficient to support the growth of Escherichia coli. J. Bacteriol. 1992;174:6682–6684. doi: 10.1128/jb.174.20.6682-6684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reuven NB, Deutscher MP. Multiple exoribonucleases are required for the 3′ processing of Escherichia coli tRNA precursors in vivo. FASEB J. 1993;7:143–148. doi: 10.1096/fasebj.7.1.8422961. [DOI] [PubMed] [Google Scholar]
- 3.Altman S, Kirsebom L, Talbot S. Recent studies of RNase P. In: Soll D, RajBhandary, editors. tRNA: Structure and Function. Washington, DC: American Society for Microbiology Press; 1995. pp. 67–78. [Google Scholar]
- 4.Li Z, Deutscher MP. Maturation pathways for E. coli tRNA precursors: a random multienzyme process in vivo. Cell. 1996;86:503–512. doi: 10.1016/s0092-8674(00)80123-3. [DOI] [PubMed] [Google Scholar]
- 5.Li Z, Deutscher MP. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, Gong X, Joshi VH, Li M. Co-evolution of tRNA 3′ trailer sequences with 3′ processing enzymes in bacteria. RNA. 2005;11:567–577. doi: 10.1261/rna.7287505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ow MC, Kushner SR. Initiation of tRNA maturation by RNase E is essential for cell viability in Escherichia coli. Genes Dev. 2002;16:1102–1115. doi: 10.1101/gad.983502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuo Y, Deutscher MP. The physiological role of RNase T can be explained by its unusual substrate specificity. J. Biol. Chem. 2002;277:29654–29661. doi: 10.1074/jbc.M204252200. [DOI] [PubMed] [Google Scholar]
- 9.Soderbom F, Svard SG, Kirsebom LA. RNase E cleavage in the 5′ leader of a tRNA precursor. J. Mol. Biol. 2005;352:22–27. doi: 10.1016/j.jmb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Mohanty BK, Kushner SR. Ribonuclease P processes polycistronic tRNA transcripts in Escherichia coli independent of ribonuclease E. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm917. doi:10.1093/nar/gkm971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDowall KJ, Hernandez RG, Lin-Chao S, Cohen SN. The ams-1 and rne-3071 temperature-sensitive mutations in the ams gene are in close proximity to each other and cause substitutions within a domain that resembles a product of the Escherichia coli rne locus. J. Bacteriol. 1993;175:4245–4249. doi: 10.1128/jb.175.13.4245-4249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wachi M, Umitsuki G, Shimizu M, Takada A, Nagai K. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5′ end of 16S rRNA. Biochem. Biophys. Res. Commun. 1999;259:483–488. doi: 10.1006/bbrc.1999.0806. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Pandit S, Deutscher MP. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 1999;18:2878–2885. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ow MC, Perwez T, Kushner SR. RNase G of Escherichia coli exhibits only limited functional overlap with its essential homologue, RNase E. Mol. Microbiol. 2003;49:607–622. doi: 10.1046/j.1365-2958.2003.03587.x. [DOI] [PubMed] [Google Scholar]
- 15.Schiffer S, Rosch S, Marchfelder A. Assigning a function to a conserved group of proteins: the tRNA 3′processing enzymes. EMBO J. 2002;21:2769–2677. doi: 10.1093/emboj/21.11.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic Acids Res. 2004;32:255–262. doi: 10.1093/nar/gkh182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schierling K, Rosch S, Rupprecht R, Schiffer S, Marchfelder A. tRNA 3′ end maturation in archaea has eukaryotic features: the RNase Z from Haloferax volcanii. J. Mol. Biol. 2002;316:895–902. doi: 10.1006/jmbi.2001.5395. [DOI] [PubMed] [Google Scholar]
- 18.Pellegrini O, Nezzar J, Marchfelder A, Putzer H, Condon C. Endonucleolytic processing of CCA-less tRNA precursors by RNase E in Bacillus subtilis. EMBO J. 2003;22:4534–4543. doi: 10.1093/emboj/cdg435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blattner FR, Plunkett G., III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, et al. The complete sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 20.Perwez T, Kushner SR. RNase Z in Escherichia coli plays a significant role in mRNA decay. Mol. Microbiol. 2006;60:723–737. doi: 10.1111/j.1365-2958.2006.05124.x. [DOI] [PubMed] [Google Scholar]
- 21.Otsuka Y, Yonesaki T. A novel endoribonuclease, RNase LS, in Escherichia coli. Genetics. 2005;169:13–20. doi: 10.1534/genetics.104.033290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schedl P, Primakoff P. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc. Natl Acad. Sci. USA. 1973;70:2091–2095. doi: 10.1073/pnas.70.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ono M, Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of mRNA. J. Mol. Biol. 1979;129:343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- 24.Arraiano CM, Yancey SD, Kushner SR. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J. Bacteriol. 1988;170:4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokobori S-I, Paabo S. Transfer RNA editing in land snail mitochondria. Proc. Natl Acad. Sci. USA. 1995;92:10432–10435. doi: 10.1073/pnas.92.22.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aiso T, Yoshida H, Wada A, Ohki R. Modulation of mRNA stability participates in stationary-phase-specific expression of ribosome modulation factor. J. Bacteriol. 2005;187:1951–1958. doi: 10.1128/JB.187.6.1951-1958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bensing BA, Meyer BJ, Dunny GM. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induce plasmid transfer system of Enterococcus faecalis. Proc. Natl Acad. Sci. USA. 1996;93:7794–7799. doi: 10.1073/pnas.93.15.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bensing BA, Sullam PM. An accessory sec locus of Streptococcuc gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 2002;44:1081–1094. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 30.Nishiyama K, Tokuda H. Genes coding for SecG and Leu2-tRNA form an operon to give an unusual RNA comprising mRNA and a tRNA precursor. Biochim. Biophys. Acta. 2005;1729:166–173. doi: 10.1016/j.bbaexp.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Altman S. A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc. Natl Acad. Sci. USA. 2003;100:13213–13218. doi: 10.1073/pnas.2235589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ow MC, Liu Q, Kushner SR. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol. Microbiol. 2000;38:854–866. doi: 10.1046/j.1365-2958.2000.02186.x. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Cole K, Altman S. The effect of a single, temperature-sensitive mutation on global gene expression in Escherichia coli. RNA. 2003;9:518–532. doi: 10.1261/rna.2198203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briant DJ, Hankins JS, Cook MA, Mackie GA. The quartenary structure of RNase G from Escherichia coli. Mol. Microbiol. 2003;50:1381–1390. doi: 10.1046/j.1365-2958.2003.03775.x. [DOI] [PubMed] [Google Scholar]
- 35.Lin-Chao S, Wong T.-T, McDowall KJ, Cohen SN. Effects of nucleotide sequence on the specificity of rne-dependent and RNase E-mediated cleavages of RNA I encoded by the pBR322 plasmid. J. Biol. Chem. 1994;269:10797–10803. [PubMed] [Google Scholar]
- 36.McDowall KJ, Lin-Chao S, Cohen SN. A + U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 1994;269:10790–10796. [PubMed] [Google Scholar]
- 37.Kaberdin VR. Probing the substrate specificity of Escherichia coli RNase E using a novel oligonucleotide-based assay. Nucleic Acids Res. 2003;31:4710–4716. doi: 10.1093/nar/gkg690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]