Abstract
Approximately 30% of human tumors characterized to date express DNA polymerase beta (pol β) variant proteins. Two of the polymerase beta cancer-associated variants are sequence-specific mutators, and one of them binds to DNA but has no polymerase activity. The Leu22Pro (L22P) DNA polymerase beta variant was identified in a gastric carcinoma. Leu22 resides within the 8 kDa amino terminal domain of DNA polymerase beta, which exhibits dRP lyase activity. This domain catalyzes the removal of deoxyribose phosphate during short patch base excision repair. We show that this cancer-associated variant has very little dRP lyase activity but retains its polymerase activity. Although residue 22 has no direct contact with the DNA, we report here that the L22P variant has reduced DNA-binding affinity. The L22P variant protein is deficient in base excision repair. Molecular dynamics calculations suggest that alteration of Leu22 to Pro changes the local packing, the loop connecting helices 1 and 2 and the overall juxtaposition of the helices within the N-terminal domain. This in turn affects the shape of the binding pocket that is required for efficient dRP lyase catalysis.
INTRODUCTION
In a mammalian cellular environment, DNA is under a continuous threat of spontaneous or chemical induced damage. Most of the time the damage is repaired (1). One of the most important repair systems is base excision repair (BER). It is estimated that at least 20 000 lesions/cell/day are repaired by BER (2,3) suggesting that this system is important for maintaining genomic integrity. Aberrant BER is likely to be associated with human cancer.
The BER pathway is important for maintaining genomic integrity. BER removes oxidative and alkylation-induced DNA damage and restores the DNA to its original form (2,3). Short patch BER results in the repair synthesis of a 1–6 nt gap whereas a gap >6 nt is filled in during long patch BER. Though polymerase beta (pol β) can participate in both pathways, its role is best characterized in short patch BER (4,5). The very first step of short patch BER is damage recognition by a DNA glycosylase. The type of glycosylase that removes the base damage usually determines the subtype of BER pathway used by the cell (6). Once the base damage is recognized and removed by a monofunctional DNA glycosylase, the product of which is an AP site, the AP endonuclease, APE1, incises the backbone and usually leaves a 5′-deoxyribose-phosphate (5′dRP) moiety. Pol β then fills in the resulting gap and removes the 5′dRP (7). However, oxidative base damage is usually removed by a bifunctional DNA glycosylase (8), which results in 3′-termini that cannot be ligated. When a 3′-dRP group is present as would likely be the case following the action of the OGG1 DNA glycosylase, the 3'diesterase activity of APE 1 removes the dRP, leaving a 3′OH (9). However, BER appears to be independent of APE1 when the base damage is removed by the NEIL DNA glycosylase (10). In this case, polynucleotide kinase (PNK) removes the 3′ phosphate that is a product of the NEIL glycoslyase. Pol β also fills in the gap after removal of oxidative damage, but it is not needed for dRP removal. After completion of gap filling by pol β, DNA ligase seals the nick.
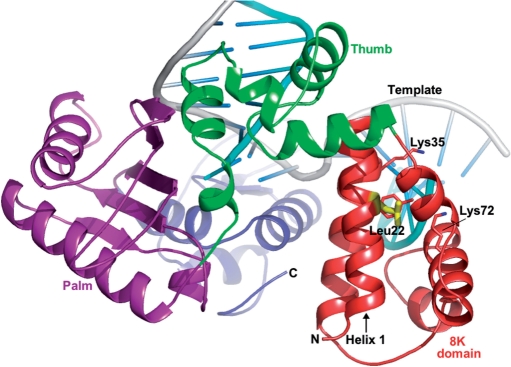
The dRP lyase activity resides within the 8 kDa amino terminal domain of pol β, as shown in Figure 1. The 8 kDa domain consists of antiparallel pairs of four helices, namely helix-1 (15–26), helix-2 (36–47), helix-3 (56–61) and helix-4 (69–78). The NMR structure shows that the pairs of helices pack with a V-like shape (11) and the N-terminus of helix-1 and the C-terminus of helix-4 are near each other. The amino acid residues that are critical for dRP lyase activity include Lys72 (helix-4), Tyr39 (helix-2) and Lys35 (12). It has been shown that the overall rate of BER is dependent upon the dRP lyase activity of pol β and not on polymerase (7).
Figure 1.
A ribbon representation of polymerase beta. The structure of pol β is depicted in domain colors showing the dRP lyase domain (8 kDa domain) in red, the N-terminal/thumb domain in green, the palm domain in magenta, the C-terminal/fingers domain in blue, the DNA template in gray and the downstream and primer strands in cyan. Leucine 22, shown in yellow, is located ∼11 Å away from the dRP lyase catalytic site (Lys72). Critical dRP lyase active site residues (Lys72, Lys35 and Tyr39) are shown in atom colors (carbon, red; nitrogen, blue).
A recent review indicates that out of 189 tumors studied, 30% of them express pol β variant proteins (13). We have shown that three of these pol β tumor-associated variants induce cellular transformation (14–16). The K289M colon and I260M prostate carcinoma variants are sequence-specific mutator polymerases that are likely to induce mutations within key growth control genes, leading to cellular transformation. The E295K gastric carcinoma-associated variant has no polymerase activity, which results in unfilled gaps that lead to genomic instability associated with cellular transformation (16). Here, we report on our studies of the Leu22Pro (L22P) gastric cancer-associated variant of pol β. We show that this variant has DNA polymerase activity but lacks dRP lyase activity. We also show that the L22P variant is unable to support BER in vitro and in vivo.
MATERIALS AND METHODS
Materials
Ultrapure deoxynucleoside triphosphates, ATP and [-γ32P] ATP (>6000 Ci/mmol, 150 mCi/ml), [-α32P]ATP (>3000 Ci/mmol) were purchased from New England Biolabs, Sigma, and Amersham Biosciences, respectively. Uracil DNA [Glycosylase (UDG) (M0280S), human AP endonuclease I (APE1) (M0282S), terminal transferase (M0252S), T4 PNK (M0201S)] and T4 DNA ligase (M0202S) were purchased from New England Biolabs. Sodium borohydride (452882) was purchased from Sigma-Aldrich.
Cloning, expression and purification of the L22P variant of pol β
The variant was generated by the Stratagene Quick-Change Site-Directed Mutagenesis kit according to the protocol of the manufacturer using pET28a-WT as a template, followed by DNA sequencing at the WM Keck facility at Yale University School of Medicine. WT and variant L22P were overexpressed in Escherichia coli strain BL21 (DE3) and purified as described previously (17).
Preparation of DNA substrates
Oligonucleotides were synthesized by WM Keck facility at Yale University. The substrates used are shown in Table 1. CII5bp, CIIR and 45AG were used for primer extension and burst kinetics, respectively. For gel shift analysis 45AG, 45AGR, 45AG1 bpDNP (45AG1 bpDNP has same sequence as 45AG except downstream DNA is not phosphorylated) were used. The primer oligonucleotide was labeled at the 5′-end by using T4 PNK and γ-32P ATP. Other oligonucleotides were 5′-end-phosphorylated with the kinase and cold ATP. After purification by a Bio–Rad spin column to remove unincorporated dNTPs, annealing was performed by mixing phosphorylated template, radiolabeled primer and phosphorylated downstream oligos in 50 mM Tris–HCl, pH 8.0, containing 0.25 M NaCl. The mixture was incubated sequentially at 95°C (5 min), slowly cooled to 50°C (for 30 min) and 50°C (for 20 min) and immediately transferred to ice. To verify proper hybridization, the product was analyzed on an 18% native polyacrylamide gel followed by autoradiography to assess the quality of annealing. For the preparation of 45AG1 bpDNP the non-phosphorylated downstream oligo was used during annealing. Likewise, for the preparation of recessed substrates 45AGR and CIIR, only template and primer were annealed.
Table 1.
DNA substrates employed in primer extension, gel mobility shift, dRP lyase and base excision repair assays
| Substrate | Sequence |
|---|---|
| 45AG | 5′ GCCTCGCAGCCGTCCAACCAAC CAACCTCGATCCAATGCCGTCC 3′ |
| 3′ CGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGG 5′ | |
| 45AG5bp | 5′ GCCTCGCAGCCGTCCAACCAAC CTCGATCCAATGCCGTCC |
| 3′ CGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGG 5′ | |
| 45AGR | 5′ GCCTCGCAGCCGTCCAACCAAC |
| 3′ CGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGG 5′ | |
| CII5bp | 5′ TTGCGACTTATCAACGCCCACAT GCTGTCTTCTCAGTTTC 3′ |
| 3′ AACGCTGAATAGTTGCGGGTGTAGTCATCGACAGAAGAGTCAAAG 5′ | |
| CIIR | 5′ TTGCGACTTATCAACGCCCACAT |
| 3′ AACGCTGAATAGTTGCGGGTGTAGTCATCGACAGAAGAGTCAAAG 5′ | |
| LPSD | 5′ CTGCAGCTGATGCGCUGTACGGATCCCCGGGTAC 3′ |
| 3′ GACGTCGACTACGCGGCATGCCTAGGGGCCCATG 5′ |
The template base or first template base is underlined in the single nucleotide gapped and recessed substrates, respectively.
In vitro primer extension assay
Primer extension experiments were conducted in a solution containing 50 mM Tris–Cl buffer (pH 8.0) 10 mM MgCl2, 2 mM DTT, 20 mM NaCl and 10% glycerol, and 50 μM each of dATP, dCTP, dGTP and dTTP. The DNA to enzyme ratio was 1:15 (50 nM: 750 nM). Reactions were carried out at 37°C for 2–60 min, after which they were stopped by addition of an equal volume of 90% formamide dye and 0.3 M EDTA. Samples were resolved by electrophoresis on 20% polyacrylamide gels containing 8 M urea, visualized and quantified using a phorphorimager.
Pre-steady-state burst experiments
A pre-steady-state burst experiment was carried out as described (17). Briefly, the radiolabeled gapped DNA (300 nM 45AG) was in 3-fold excess relative to pol β (100 nM). Also, a modified burst reaction was carried out for L22P where the DNA to polymerase ratio was 18 μM:6 μM. The burst experiment was performed at saturating concentrations of dTTP while minimizing any substrate inhibition, which may occur with excess dTTP. Reactions were initiated by rapid mixing of the pol β-DNA and Mg-dTTP solutions. At selected time intervals, the reactions were quenched with 0.3 M EDTA. The reaction products were separated on a 20% denaturing polyacrylamide gel. Data from the burst experiments were fitted to the burst equation: [product] = A(1 − exp(−kobst) + kSSt), where A is the amplitude of the burst, kobs is observed first-order rate constant for dTTP incorporation and kss is the observed linear rate constant.
Gel mobility shift assay
Various concentrations of WT pol β protein (0.1–1000 nM) and L22P pol β protein (0.1–2000 nM) were incubated with 0.1 nM radiolabeled gapped DNA substrate in buffer containing 50 mM Tris–Cl, pH 8.0, 100 mM NaCl, 10 mM MgCl2, 10% glycerol, and 0.1% Nonidet P-40 at room temperature (23°C) for 15 min. Samples were loaded onto a 6% native polyacrylamide gel with the current running at 300 V at 4°C. After the sample was loaded, the voltage was reduced to 150 V. Bound protein was quantified using Imagequant software, after scanning the gel using a Molecular Dynamics Phosphorimager. Protein bound to DNA resulted in a shift of the DNA on the gel when compared to DNA without bound protein. Fraction bound is the ratio of the intensity of all shifted species divided by the total. The dissociation constant for DNA (KD) was estimated from fitting the bound protein (Y) versus protein concentration (x) with the equation: Y = [(mx)/(x + KD)] + b, where m is a scaling factor and b is the apparent minimum Y value.
Preparation of 5′dRP substrate
The 3′end of DNA substrate LPSD, which contained a single U at position 19 from 3′ end, was radiolabeled using [−α32P] ATP and terminal transferase and annealed with its complementary oligo. This DNA duplex was treated with UDG (2 U/1 pmol DNA) in 50 mM HEPES (pH 7.5) at 37°C for 10 min, followed by treatment with human AP endonuclease, APE1 (2 U/1 pmol DNA) in buffer R (10 mM MgCl2, 20 mM KCl and 2 mM dithiothreitol) at 37°C for 10 min, which incises the phosphodiester backbone on the 5′-side of the AP site and leaves a 3′-OH and a 5′-dRP residue. Due to the labile nature of the AP site, the 5′-dRP-containing DNA substrate was prepared just before use.
5′-dRP lyase assay
The assay was set up according to Prakash et al. (18) with minor modifications. Typically, reaction mixtures (24 µl) contained the DNA polymerases (200–400 nM) and the 5′-dRP-containing DNA substrate (100 nM) in buffer R [50 mM HEPES (pH 7.5), 10 mM MgCl2, 20 mM KCl and 2 mM dithiothreitol]. Reactions were incubated at 37°C for 10 min. The reaction product was stabilized by the addition of 2 M sodium borohydride to a final concentration of 340 mM, followed by incubation on ice for 30 min. Stabilized (reduced) DNA products were ethanol-precipitated in the presence of 0.1 µg/ml of tRNA, resuspended in water, and an equal volume of formamide dye was added. These products were resolved on a 20% polyacrylamide gel and visualized with a Storm 860 PhosphorImager (Molecular Dynamics, Inc.).
Trapping of polymerase–DNA complexes
To capture the trapped complex of DNA with pol β, 400 nM enzyme (WT or L22P) was mixed on ice in buffer R with DNA substrate containing a 5′-dRP residue. This was followed by immediate addition of 20 mM sodium borohydride solution. The tubes were kept on ice for another 30 min to stabilize the complex. After removal of the dRP moiety, the intensity of the trapped substrate will be reduced. To determine whether the enzyme dissociated from the DNA, it was first pre-incubated with the DNA substrate at 37°C for 10 and 30 min before the addition of sodium borohydride. The tubes were immediately transferred to ice after addition of the borohydride for 30 min to stabilize the complex. The reactions were terminated by the addition of sodium dodecyl sulfate (SDS)-containing loading buffer, and the cross-linked polymerase-3′-32P-labeled DNA complexes were resolved on a 10% SDS–polyacrylamide gel. The products were visualized by autoradiography.
BER assay
As a BER substrate, 5′-end labeled LPSD substrate (Table 1) was used. For reconstituted BER with purified proteins, we followed the method as described by Prakash et al. (18) with some minor modifications. Typically, 100 nM UDG-treated substrate was first incubated for 10 min with commercially available APE1. Approximately 50 nM APE-treated substrate was incubated with purified pol β (200–10 000 nM) in BER buffer (45 mM HEPES pH 7.8, 70 mM KCl, 2 mMDTT, 7.5 mM MgCl2, 0.5 mM EDTA, 2 mMATP and 20 μM each of dATP, dTTP, dCTP and dGTP) for another 10 min at 37°C with or without T4 DNA ligase. Finally, EDTA containing formamide dye was added to stop the reaction. The repaired product was resolved on a 20% denaturing polyacrylamide gel followed by visualization on the Phosphorimager.
Methylmethane sulfonate (MMS) sensitivity assay
MEF cell line 88TAg (pol β−/−) (19), was maintained in Dulbecco modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), l-glutamine (Invitrogen), β-mercaptoethanol (Sigma) and penicillin–streptomycin (Invitrogen) at 37°C in a humidified 5% CO2 incubator. The GP2-293 cell line (Clontech) was used for retroviral packaging.
The pRVY-Tet plasmid expressing WT pol β was described previously (14). Briefly, in this vector, the left-hand retroviral long terminal repeat drives expression of tTA tetracycline (Tet) transactivator, the tetO/CMV promoter drives expression of the pol β protein in a Tet-repressible manner and an internal SV40 early promoter drives expression of the hygromycin resistance gene. Thus, when Tet is present in the growth medium, expression of the WT or L22P proteins is turned off. However, expression of these proteins occurs when Tet is removed from the growth medium. The L22P variant was constructed by site-directed mutagenesis of pRVY-Tet containing WT pol β, using a kit (Stratagene).
For retroviral packaging each plasmid was cotransfected into GP2-293 cells along with equal amount of pVSVG DNA, using FuGene 7 (Roche). Virus was collected in 72 h. Stably transfected GP2-293 cells were selected in presence of 300 µg/ml hygromycin B (HygB; Invitrogen) and used for preparation of high titer virus. Retroviral transduction of 88-TAg cells was performed using a standard protocol (Clontech). Use of high titer virus insured at least 50% efficiency of infection. Infected cells were grown in presence of 300 µg/ml hygromycin B until confluent. Cells were split once at 1:20 dilution and this passage was designated as passage 1. Non-infected cells were used as a negative control for drug selection and displayed 100% lethality by passage 1.
Recombinant cell lines were grown no more than six passages in normal growth medium containing 300 µg/ml hygromycin B. Western blotting was used as described (16) to determine if the WT and L22P variants were expressed in the absence of Tet.
To test the sensitivity of the cells to MMS, we used the Cell Titer Assay (Promega) according to manufacturer's recommendations. Briefly, cells were seeded at 1000 cells/well and left to attach overnight. The next day the cells were treated for 1 h with various concentrations of MMS in normal growth medium. The medium was then changed and the cells were incubated for 72 h under normal growth conditions. Twenty microliters of the cell titer solution was added into each well and the OD490 was measured by a Spectra Max plate reader. A non-treated control for each cell line was counted as 100% after background subtraction. Each cell line was tested in quadruplicate. Each experiment was repeated at least four times using independently created cell lines. Results were analyzed by ‘Prism’ software (GraphPad). The significance of differences in sensitivity that we obtained were assessed by a non-parametric test and accepted if P <0.05.
Modeling and molecular dynamics
The programs O, NAMD2, VMD (20–22) were used to introduce and analyze the mutation at position 22 in pol β. The models of native and variant pol β are based on a DNA co-crystal structure (PDB: 1mq3). The N-terminal domain was subjected to a periodic-boundary equilibration followed by a 10 ns molecular dynamics simulation. The calculations were performed using the Non-equilibrium Atomic Molecular Dynamics 2 (NAMD2) simulation package version 2.5. For the simulations the models were restricted to a domain comprising residues 1–118. The Charmm27 force field (23) was used to model the intramolecular interactions within the N-terminal domain. The TIP3P model was used in the description of the water molecules. The N-terminal domains were placed in a sphere (non-periodic boundary) of water molecules with a 36 Å radius exceeding the recommended NAMD2 default sphere radius by more than 10%. (21). Long-range non-bonding terms were calculated with a 12 Å cutoff for electrostatic and van der Waals interactions. All hydrogen bond lengths were held constant with the SHAKE-RATTLE-ROLL algorithm. The simulations were carried out at a temperature of 310 K. The simulation temperature was maintained using a Langevin thermostat with a coupling constant of 5 ps−1. The lengths of the simulations were determined by the proper convergence of the monitored properties (both molecules ran for a period of 20 ns simulations). The time step for all simulations was set to 2 fs. Trajectory analysis and molecular graphics images were generated using VMD. The time evolution of the radius of gyration (Rgyr), alpha-carbon alpha-carbon distances and hydrogen-bonding interactions were monitored during the simulation. Root mean-square deviation analyses (RMSD) were performed to evaluate systems mobility and proper convergence (data not shown). Ramachandran maps and scatter plot diagrams were generated to document changes in the overall structure of the N-terminal domain over time.
RESULTS
L22P is a less active polymerase than wild-type pol β
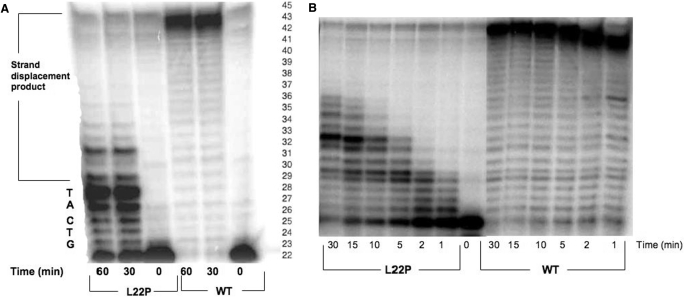
To determine if L22P had polymerase activity, a primer extension assay was carried out with 5 bp gapped DNA substrate (CII5bp) to assess the polymerase activity of the L22P variant in comparison to WT. Our data in Figure 2A show that both WT and L22P fill the gap. However, under our assay conditions, WT pol β exhibits robust strand displacement synthesis whereas L22P appears to be quite distributive. We performed an additional primer extension experiment with recessed DNA substrate (CIIR). As shown in Figure 2B, L22P extends the primer much less efficiently than WT. Thus, the L22P variant is less active than WT pol β.
Figure 2.
L22P is a less active polymerase than WT pol β in primer extension assay. (A) Gapped DNA substrate. Purified WT and L22P enzymes (750 nM) were incubated with the radiolabeled 5 bp gapped DNA substrate (50 nM) (CII5bp) and all four dNTPs for 30 min and 1 hour at 37°C as described in Materials and Methods section. The sequence of the template within the gap is indicated on the left side of the Figure and the right side represents the number of the bases from the position of the primer. (B) Recessed DNA substrate. Purified 750 nM WT and L22P enzymes were incubated with the 50 nM radiolabeled recessed DNA substrate (CIIR) and all four dNTPs for indicated times at 37°C as described in Materials and Methods section. Products were separated by denaturing gel electrophoresis and visualized using a Phosphorimager.
L22P has lower affinity for gapped DNA
The slower rate of DNA synthesis by L22P could be due to a lower DNA-binding affinity than WT. To assess the affinity of L22P for DNA, we carried out gel mobility shift assays of WT and L22P using three different substrates: 1 bp gapped (45AG), recessed (45AGR) and 1 bp gap but without a 5′ phosphate on the downstream DNA (45AG1bDNP). Irrespective of the substrates utilized, L22P has lower affinity for DNA than WT pol β, as shown in Table 2. This effect is much more pronounced in the case of the 1 nt gapped DNA, with which L22P has 19-fold lower affinity than WT.
Table 2.
L22P has lower DNA-binding affinity than WT
| Polymerase β | KD (DNA), nM | ||
|---|---|---|---|
| 45AGR (Recessed) | 45AGa (1-bp gapped) | 45AG1bpDNPb (1-bp gapped) | |
| WT | 84 ± 18 | 12 ± 2 | 20 ± 3 |
| L22P | 455 ± 100 | 229 ± 50 | 291 ± 45 |
aDownstream oligonucleotide is phosphorylated at 5′ end.
bDownstream oligonucleotide is not phosphorylated.
L22P does not exhibit burst kinetics under usual reaction conditions
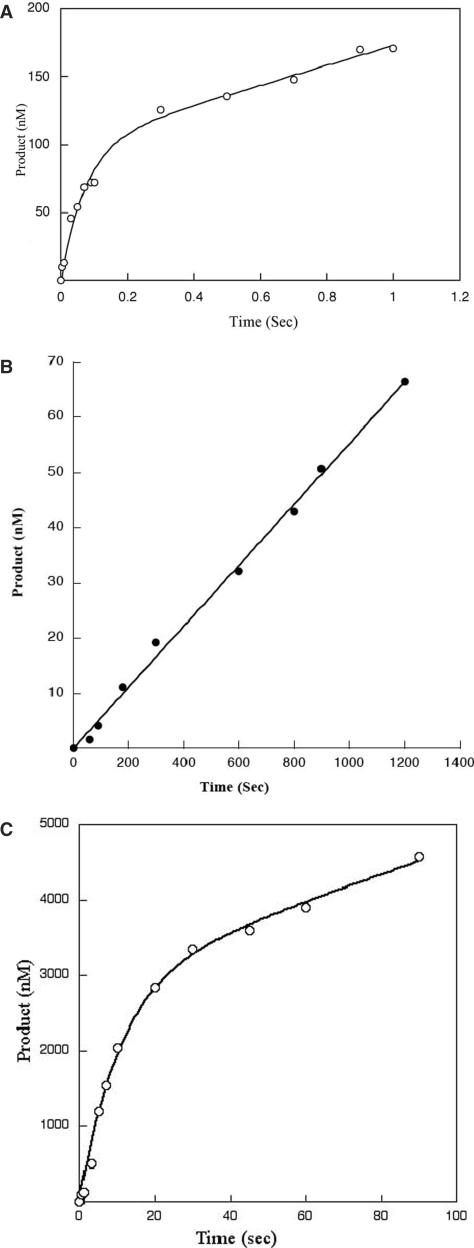
Next, to determine if L22P had a rate-limiting step that was similar to that of WT pol β, we measured the reaction rate of L22P in a pre-steady-state kinetic assay. We used 300 nM radiolabeled single-nucleotide gapped DNA substrate, and 100 nM pol β. A typical biphasic burst curve was observed for WT, with a burst rate of 12 s−1, as shown in Figure 3A. However, no product formation was detected for the L22P variant within the maximum time limitations of our experiment, which was 3 s. We then repeated this experiment with L22P for longer amounts of time (0–1400 s). Instead of a biphasic curve of product formation we obtained a slow linear rate of 0.06 s−1 for L22P, as shown in Figure 3B. This suggests that the rate-limiting step of L22P is different from that of WT. Considering the KD (DNA) obtained from the gel shift assay, the percent bound can be estimated by the equation% bound = [E]/[E] + KD, where E is the enzyme concentration used in the experiment. So percentage bound will be 90% and 43% for WT and L22P, respectively, under our pre-steady state experimental conditions.
Figure 3.
L22P shows no burst of product formation under usual reaction conditions. A pre-incubated solution of WT or L22P and single-nucleotide gapped DNA was mixed with a solution of 100 µM dTTP containing 10 mM MgCl2. The reactions were terminated by EDTA, and the products were resolved by denaturing sequencing gel electrophoresis. (A) Insertion of dTTP into a single-nucleotide 300 nM gapped DNA substrate by 100 nM WT (45AG) was measured using a chemical quench-flow apparatus at 37°C. Data for WT were fit to the burst equation with a kobs of 12.0 ± 1.6 s−1 and a steady-state rate constant of 1.1 s−1. (B) Insertion of dTTP into a 300 nM single-nucleotide gapped DNA substrate (45AG) by 100 nM L22P was measured manually at 37°C at various time points as indicated on the x-axis. The data were fit to the linear equation with a steady-state rate constant of 0.06 ± 001 s−1. (C) Insertion of dTTP into a single-nucleotide 18 μM gapped DNA substrate (45AG) by 6 μM L22P was measured using chemical quench-flow apparatus at 37°C at various time points as indicated on the x-axis. The data were fit to the burst equation with a burst rate of 0.09 ± 01 s−1 and a steady-state rate constant of 0.006 s−1.
To determine the burst rate under conditions in which L22P would be more than 90% bound, we used 6 μM enzyme and 18 μM DNA and repeated the experiment. Under these conditions, L22P showed a typical biphasic curve (Figure 3C) with a very slow burst rate of 0.09 ± 0.01 s−1and a steady-state rate of 0.006 s−1. Because the protein concentration was so high, we suggest that L22P may not be acting as an enzyme and that product formation may result from molecular crowding of DNA and protein.
L22P has no dRP lyase activity
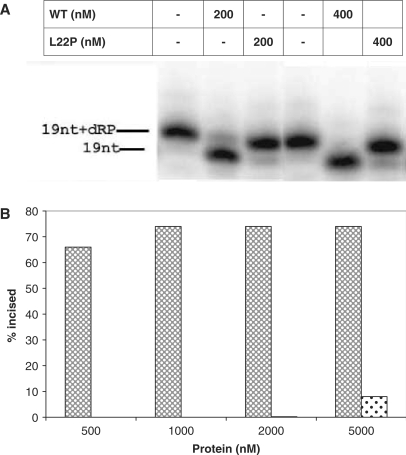
Leu22 maps within the 8 kDa domain, which contains the catalytic site of the dRP lyase of pol β, so we explored the idea that altering this residue to Pro would have an effect on the dRP lyase activity. We performed the dRP lyase assay at two different protein concentrations (200 and 400 nM), as described in the legend to Figure 4A. As shown in Figure 4A, WT removes the dRP group very efficiently as suggested by the presence of 19 nt lower band while L22P is devoid of dRP lyase activity in our reaction conditions.
Figure 4.
L22P lacks dRP lyase activity. The 5′-dRP-containing DNA substrate, LPSD (100 nM), was incubated in buffer R at 37°C for 10 min with increasing concentrations of pol β as indicated in the figure, followed by the addition of 340 mM NaBH4 and stabilization of the reaction product as indicated in Materials and Methods section. The products were analyzed by a denaturing 16% polyacrlyamide gel and visualized by autoradiography. (A) Image of the denaturing gel. (B) A graphical representation of the percentage of incised product versus protein concentration. The cross-hatched bar represents WT and the dotted bar represents L22P.
To determine whether the lack of dRP lyase activity results from the low DNA-binding affinity of L22P, we carried out a dRP lyase assay in which 90% of L22P is bound to DNA. We observed a negligible amount of dRP lyase activity under these conditions. Under conditions in which >95% of L22P is bound to DNA, (5000 nM) we did observe a small amount of incised product, as shown in Figure 4B. These results suggest that Leu22 is important for the dRP lyase activity of pol β.
L22P forms a Schiff's base complex
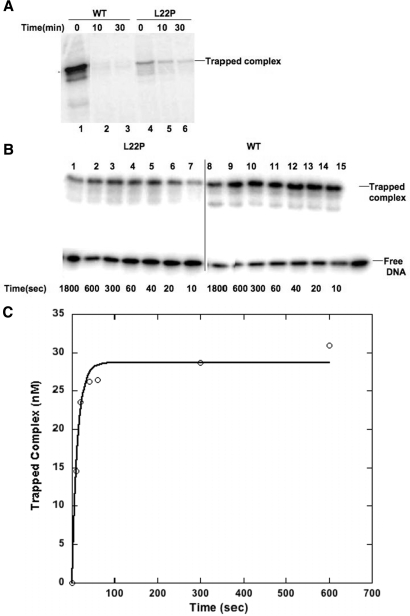
There is no evidence that Leu22 has a direct role in the catalysis of dRP group removal. However, our data indicate that altering Leu to Pro nearly abolishes the dRP lyase activity of pol β. Previous reports suggest that the dRP lyase reaction is catalyzed by β-elimination after Schiff's base formation (24). The Schiff's base complex is formed between Lys72 and the DNA. This complex is normally very transient but can be trapped using sodium borohydride as a reducing agent (18). We carried out a trapping experiment to determine whether the L22P variant is capable of forming a Schiff's base. As shown in Figure 5A (lane 4), L22P forms a Schiff's base complex, the amount of which appears to be much less than WT. After Schiff's base formation, the reaction follows a β-elimination reaction followed by phosphate removal. Thus, the intensity of the trapped polymerase and DNA complex should diminish if borohydride is added after the reaction is over. For WT pol β, we observed this reduction of trapped product formation (Figure 5A, lanes 2 and 3), suggesting there is no dRP lyase substrate remaining at the end of the reaction. In contrast, a significant amount of substrate remains in the case of L22P (lanes 5 and 6). This is also observed when the concentration of L22P is 5000 nM (L22P is >90% bound to DNA) and that of DNA is 100 nM (data not shown). These results are consistent with the interpretation that L22P does not possess robust dRP lyase activity and that this lack of activity is not due to limited DNA binding.
Figure 5.
(A) Trapping of Schiff's base complexes. Trapping of Schiff's base complexes of WT and L22P before and after the dRP lyase reaction. A total of 400 nM WT (lanes 1–3) and 400 nM L22P (lanes 4–6) were mixed with the 100 nM 5′-dRP-containing 3′-32P-labeled LPSD DNA substrate on ice, followed by incubation at 37°C for 0 (lanes 1 and 4), 10 (lanes 2 and 5) or 30 min (lanes 3 and 6). A solution of 20 mM NaBH4 was added and the reaction was incubated further on ice for 30 min to stabilize the product. After the addition of SDS-containing loading buffer, the samples were resolved on a 10% SDS–polyacrylamide gel, and the trapped polymerase–DNA products were analyzed by autoradiography. (B) Trapping of Schiff's base complex formation in the presence of sodium borohydride. A total of 5000 nM L22P (lanes 1–7) and 500 nM WT (lanes 8–14) were mixed with 50 nM 5′-dRP-containing 3′-32P-labeled LPSD DNA substrate on ice in continuous presence of 20 mM NaBH4. At each time interval (10–1800 s), 20 μl of reaction mix was taken out and added to equal volume of SDS gel-loading dye to stop the reactions. The samples were resolved on a 10% SDS–polyacrylamide gel, and the trapped polymerase–DNA products were analyzed by autoradiography. (C) Determination of rate of Schiff's base formation by L22P. Amount of trapped Schiff's base complex by L22P was plotted against time. The data were fit to the single exponential equation with a rate of 0.074 ± 0.009 s−1.
Since we observed a reduced amount of Schiff's base formation by L22P (Figure 5A, lane 4), we were interested to know the rate of the reaction. We determined the rate of Schiff's base formation (Figure 5B) for WT and L22P at conditions where both of the proteins are 95% bound to DNA. As shown in Figure 5B (lanes 8–14), at 500 nM concentration, WT forms a saturating amount of Schiff's base complex very quickly (within 10 s) and we were not able to calculate the reaction rate. For L22P, even at 5000 nM concentration, the rate of Schiff's base formation is slow. We plotted the product of trapped complex versus time, which fit the single exponential equation and yielded an apparent rate of Schiff's base formation of 0.074 s−1 (Figure 5C).
L22P cannot support BER
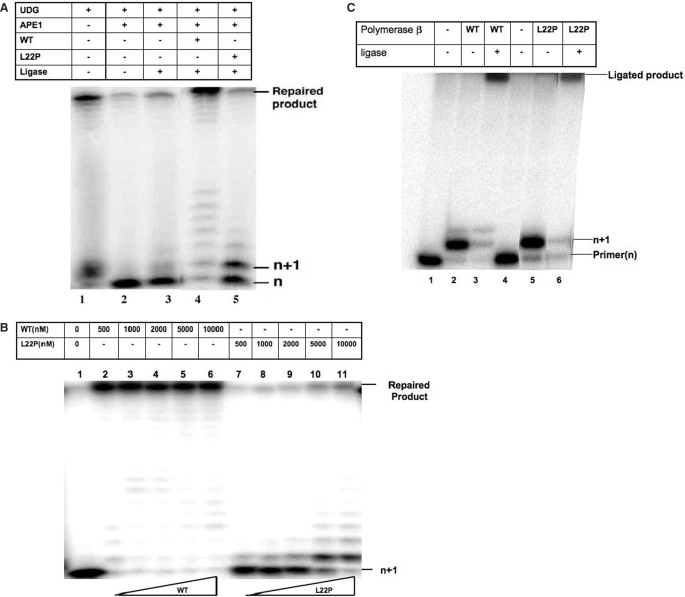
Since the primary activity of pol β is to function in the repair of small single-base damage in DNA, we tested the L22P variant's ability to participate in short patch BER. An in vitro BER assay was carried out using 5′end-labeled LPSD substrate (Table 1) that contains a uracil, and purified proteins as described in detail in the Materials and Methods section. Lane 1 shows the DNA substrate after the uracil has been removed by UDG. After cleavage of the backbone with APE1 (lane 2) the product cannot be ligated by T4 DNA ligase (lane 3). However, addition of 400 nM WT pol β and T4 DNA ligase to the reaction mixture results in nearly complete BER as shown in Figure 6A (lane 4). In contrast, when L22P is added to the reaction, the n + 1 product appears, which suggests L22P has polymerase activity. dRP group removal is necessary prior to ligation but the L22P variant has no dRP lyase activity. Therefore, we did not observe any ligated product (Figure 6A, lane 5) in presence of L22P.
Figure 6.
L22P does not support BER.(A) Reconstituted BER with purified proteins. Lane 1, annealed oligo substrate, treated with uracil DNA glycosylase (UDG); lane 2, UDG-treated substrate incubated with APE1 for 10 min; lane 3, UDG treated substrate incubated with APE1 and T4 DNA ligase for 10 min; lane 4, UDG-treated substrate, incubated with APE1, 400 nM of purified WT pol β and T4 DNA ligase for 10 min; lane 5, UDG-treated substrate, incubated with APE1, 400 nM L22P pol β and T4 DNA ligase for 10 min. (B) L22P lacks BER activity even at high concentrations. A reconstituted BER assay was carried with increasing protein concentrations (500–10 000 nM). Lane 1: UDG- and APE1-treated substrate, lanes 2–6: BER assay with WT, lanes 7–11: BER assay with L22P. (C) L22P can fill in a single nucleotide gap. A single-nucleotide primer extension assay was carried out in presence of 50 μM dTTP and 10 mM MgCl2 using 45AG (50 nM) as substrate; 500 nM WT and 5000 nM L22P were used to carry out the reaction at 37°C for 10 min. Reactions were performed in presence (lanes 3 and 6) and absence (lanes 2 and 5) of T4 DNA ligase.
To confirm that the defect in BER is not due to limited DNA-binding affinity of L22P we repeated the reconstituted BER assay with a range of higher protein concentrations (500—1000 nM) (Figure 6B). WT completed repair at all concentrations tested (lanes 2–6). At increasing concentrations of L22P we observe an increase in n + 1 product formation (lanes 7–11), which suggests that L22P is capable of DNA synthesis. At 5 and 10 μM concentration, where L22P is likely greater than 95% bound, we observed formation of the maximum amount of n + 1 band but there was very little ligated product formed (∼5%) (lanes 10 and 11). These data suggest that repair appears to be hindered by L22P's inability or reduced ability to remove the dRP moiety.
Bifunctional DNA glycosylases usually function in the repair of various oxidized bases, and they remove the damaged base and leave a modified 3′end and a 5′-phosphate. In this case, pol β would be required to fill in the gap but not to remove the 5′dRP group. Therefore, we asked if the L22P variant could fill in a single nucleotide gap with a 3′-OH and a 5′-phosphate that could then go on to be ligated. As shown in Figure 6C, both WT and L22P are able to fill in a single nucleotide gap, which can then be ligated by T4 DNA ligase.
Molecular modeling results
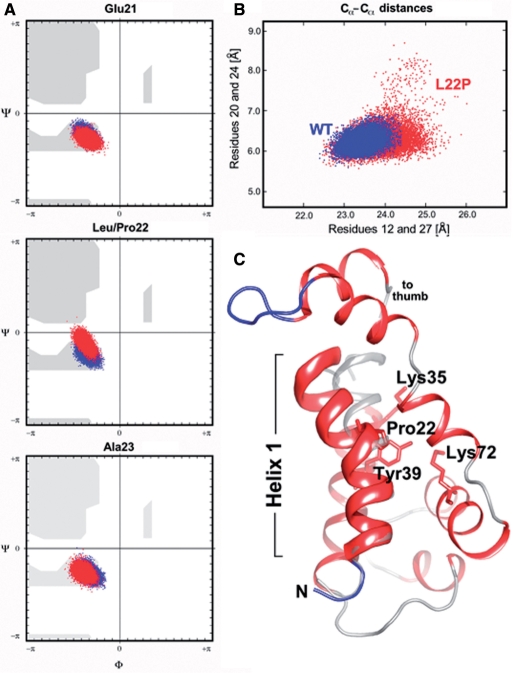
In order to provide a structural framework for understanding the characteristics of the L22P variant, MD calculations of pol β's N-terminal domain (comprising residues 10–118 of the 8K and part of the thumb domain) of both wild-type and the L22P variant were carried out for a total of 10 ns to ensure the simulations converge. During the simulations the radius of gyration (Rgyr) of the L22P variant increased by ∼3% as compared to the wild-type structure (data not shown). Upon completion of the MD calculations the N-terminal domains of the native structure and the L22P variant show backbone atom RMS deviations to the starting model (PDB code: 1MQ3) of 2.17 and 2.41 Å, respectively. A maximum RMS deviation of 2.7 Å was observed at 15.9 ns coinciding with a dramatic bend in helix 1 (Figure 7C). The conformational changes of the L22P N-terminal domain suggest that the local structure in helix 1 and the packing of side-chains adjacent to Pro22 are significantly affected by the mutation.
Figure 7.
Analysis of MD simulations. (A) Changes in Ramachandran backbone angles for residues Glu21, Leu/Pro22, Ala23 along the 20 ns trajectory. The backbone torsion angles for the wild-type pol β; are shown in blue and the phi/psi angles of the Pro22 mutant are depicted in red. Note the deviations in backbone torsion angles in the middle panel (site of mutation; Leu22, blue dots; Pro22, red dots). (B) Scatterplot of inter-residue distances (20–24 and 21–27) The distortion of helix 1 during the dynamics simulation has been visualized by plotting the distances between residues 20–24 and 21–27 (highlighted by the dotted line in ribbon diagram below). Residues 20, 21, 24 and 27 lie in the same plane as the bend in helix 1. (C) Cartoon drawing of the 8 K domain of L22P at 15.9 ns showing a significant bending of helix 1 at residue 22. Helix 1 is highlighted as a thickribbon, the remainder of the 8 K domain is depicted as a thin ribbon. The N-terminal helix of the wild-type pol β; crystal structure (PDB code 1bpy) is overlaid in gray (semi-transparent rendering). Note that helix 1 in wild-type polβ is nearly perfectly straight.
Backbone angles and inter-residue distances within helix 1 were analyzed in detail. A selection of Ramachandran maps for residues 21–23 and inter-residue distance scatterplots calculated or residues Thr20 to Asn24 and Asn12 to Lys27 are depicted in panels A and B of Figure 7. While the peptide backbone angles for the sampled native (blue dots) and L22P variant (red dots) structures are in good agreement as shown for Glu21 and Ala23, they clearly deviate at position 22, the site of mutation (Figure 7A). The proline residue apparently introduces a much greater degree of flexibility into helix 1 as seen from the scatter plot in Figure 7B. For the variant (red dots) the distances between the alpha carbon atoms of Thr20 and Asn24 and Asn12 and Lys27 vary more widely than the corresponding distances sampled for the native structure (blue dots). Especially, the residues in the close vicinity of Pro22 (Thr20, Gln21 and Ala23, Asn24) experience a great deal of stretching and scrunching. Figure 7C highlights a structure from the L22P trajectory at around 15.9 ns where the alpha carbon distance between Asn12 and Lys27 appears to reach a global minimum indicative of a dramatic bend within helix 1. The corresponding wild-type helix, which is almost perfectly straight, is superimposed for reference. Closer inspection of superimposed intermediate structures of L22P indicate that the loop connecting helices 1 and 2 (residues 30–34) displays a great deal of variability, which is likely a direct result of introducing Pro22 into helix 1. The wild-type structures, on the other hand, appear far less dynamic in behavior with respect to that of helix 1 and the loop connecting to helix 2. In L22P, the side chains of Ser30 and His34 in particular appear to affect the position of NZ of Lys35 directly. The position of Lys35 has been shown to affect dRP lyase activity (25).
L22P does not function in BER in cells
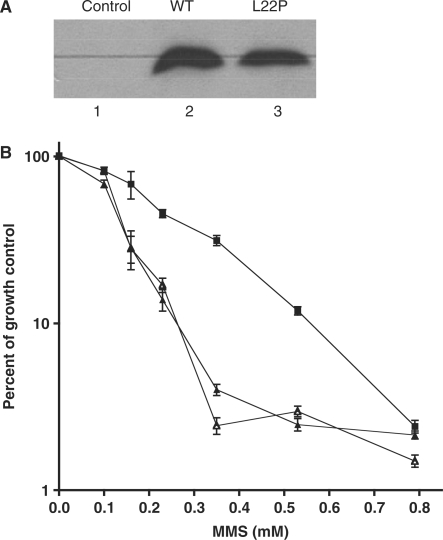
To determine if the L22P variant could function in the BER of alkylating agents in cells, we expressed this protein in pol β-deleted MEFs and characterized the survival of the cells in response to treatment with various doses of MMS. As shown in the western blot in Figure 8A, the WT and L22P proteins are expressed in the MEFs. The pol β-deleted MEFs are quite sensitive to MMS as has been shown previously (4). Expression of WT pol β in these cells confers resistance to MMS, whereas expression of L22P does not, as shown in Figure 8B. This suggests that the L22P variant is not able to function in BER in cells.
Figure 8.
Expression of L22P in MEFs sensitizes them to MMS. (A) Western blot. Lane 1, negative control; lane 2, cells with pRVY-Tet expressing WT pol β and lane 3, cells with pRVY-Tet expressing L22P. The blot was probed with monoclonal antibody to pol β as described (15). (B) Survival curves. (Open triangle), pol β-deleted MEFs; (filled square), MEFs expressing WT pol β; (outlined triangle) MEFs expressing L22P pol β.
DISCUSSION
In this study, we investigated the biochemical behavior of the L22P gastric cancer-associated variant of pol β. Our results suggest that this variant has lower affinity for DNA, significantly reduced dRP lyase activity, and that L22P cannot support BER. Molecular modeling studies support our biochemical data. Altogether, our results imply that an amino acid residue that is some 10.6 Å from the active site cleft of the dRP lyase critically affects the catalysis of dRP removal.
The L22P variant has a lowered DNA-binding affinity
The physiological substrate for pol β is likely single-nucleotide gapped DNA. In general, pol β has high affinity for DNA. This affinity is highest for 1 base gapped DNA, followed by 5–6 base gapped and recessed DNA. The presence of downstream DNA is required for efficient and tight binding. The presence of phosphorylated versus non-phosphorylated downstream DNA does not increase the affinity for DNA but increases polymerase fidelity (26,27).
The N-terminal 8 kDa domain interacts with the downstream DNA whereas the thumb subdomain acts as a primer grip. NMR studies suggest that the DNA-binding site of the 8 kDa domain is located on the C-terminal part of the HhH motif. Residues important for DNA binding in the 8 kDa domain are Lys41, Lys60, His34, Arg40, Tyr39, Lys 68, Lys 72 and Arg83 (12). Although there is no structural detail available for Leu22 regarding DNA interactions, it is known that Leu is a helix-stabilizing amino acid (28). Thus, Leu22 most likely contributes to the overall stability of the neighboring HhH. Our results suggest that alteration of the helix stabilizing residue leucine to a helix breaker proline reduces the DNA-binding affinity by several fold (19×). Thus, if helix 1 becomes destabilized, the effect is probably propagated to the DNA-binding site.
BER and the dRP lyase activity of pol β
Pol β possesses DNA polymerase and dRP lyase activities, both of which are known to be important for BER. Previous reports showed that the dRP lyase deficient K72A pol β was unable to support BER in cell extracts prepared from pol β-deleted cells and that the 8 kDa domain alone was able to complement the MMS sensitivity of these cells (29,30). The presence of the dRP group prevents DNA ligase from joining the phosphodiester backbone.
Our data demonstrate that L22P has no significant dRP lyase activity even under conditions where likely >95% of the DNA is bound by the protein. Under these conditions, we observed formation of the n + 1 band, confirming the ability of L22P to catalyze DNA synthesis, but there was very little ligated product formed (∼5%) (Figure 6B, lanes 10 and 11). The L22P variant is able to interact with DNA ligase III and XRCC1 (data not shown), ruling out the possibility that a protein interaction deficiency is responsible for its inability to support BER. These results indicate that L22P is unable to support BER initiated by UDG, because it is not able to remove the dRP group. We have also shown that L22P is not able to support BER in cells treated with MMS, likely because it is not able to remove the dRP group. It has been shown by in vitro reconstituted BER assays that are initiated with UDG, that the overall rate-limiting step of BER is the dRP lyase reaction of pol β (7). However, this is not likely to be the case when oxidized bases are removed by either OGG1 or NEIL DNA glycosylase (9,10).
Altered DNA binding and abolished dRP lyase activity are due to structural changes in the N-terminal domain
Leu22 is located in the N-terminal (8 kDa) domain of DNA polymerase beta and is not able to contact the DNA substrate directly. Previous studies have indicated that residues important for dRPase activity are Tyr39, Lys68 and Lys72 (12,31,32). A study from Wilson et al. (31) showed that altering Lys72 to Ala abolishes dRP lyase activity without changing DNA binding. Matsumoto and colleagues (24,32) discovered that the dRP lyase reaction occurs via formation of Schiff's base followed by a beta elimination reaction. Further structural analyses revealed that Lys72 is the main catalytic residue responsible for Schiff's base formation (12).
Leu22 is a hydrophobic residue located in the middle of the helix 1 (Figures 1 and 7C). Circular dichroism studies do not indicate that there are significant changes in the overall alpha helical content (data not shown) and thermal stability (melting temperature, Tm) of L22P, suggesting the effect of Pro22 on the 8 kDa domain might affect the tertiary structure or perhaps the juxtaposition of pol β subdomains, changes not tractable by circular dichroism. The side-chain volume decreases typically by ∼30% when Leu22 is replaced by Pro in the variant structure. Consequently, the changes in packing density along with the observed increase in Rgyr in the L22P variant as compared to the fully equilibrated native structure could adversely affect duplex binding and catalytic activities of the L22P mutator.
Leu22 is some 10.6 Å (Cα-C) away from the catalytic Lys72 residue. Our biochemical and computational data suggest that it can affect overall catalysis for dRP lyase activity most probably by repositioning both helix 1 and an adjacent loop (residues 30–34), which in turn affects the catalytic pocket of dRP. Several important residues (Glu26, Ser30, His34, Lys35, Tyr39, Lys68 and Lys72) form the lyase catalytic pocket (25). The first step of lyase reaction is correct positioning of the flexible sugar moiety followed by ring opening. His34 has been proposed to be involved in the base-stacking interaction with the downstream template base of gapped DNA. Protonation of Lys72 might induce ring opening. Feng et al. (33) have proposed that residue Lys35 or Lys68 could initiate the reaction by protonating the deoxyribose O-4′, leading to opening of the ring. Lys72 is then deprotonated, ready for the nucleophilic attack. Mullen et al. (11) proposed from their structural data that the deprotonated form of Lys72 is stabilized by residue Tyr39. Following this stabilization, the beta elimination reaction takes place, which could be facilitated by Glu26. The last step of the lyase reaction is the removal of dRP from the Schiff's base complex. Lys35 is suggested to participate in the stabilization of the leaving phosphate group. The rigidity of the backbone around residues Lys72, Lys35 and Tyr39 allows a well-formed active site for dRP removal between DNA and protein. The bending and the inherent flexibility of helix 1 in the L22P variant causes local and global conformational changes in the structure of the N-terminal domain that may well cause a decrease in the binding affinity for gapped duplex DNA and a dramatic reduction in the actual catalytic turnover rate of the dRP lyase activity.
Our trapping experiments suggest that the L22P variant of pol β somehow retains the ability to form a Schiff's base (Figure 5A) but the rate of Schiff base formation is slower than WT (Figure 5B). It is most likely that residues Ser30 and His34 are not placed in the right position to aid Lys72 in completing the beta elimination reaction. Our results also suggest that the final step of phosphate removal is impaired in case of L22P since we did not observe rapid phosphate removal as with WT, indicating that Lys35 has most probably lost its rigidity that is required for correct active site geometry.
L22P and cancer
Our results showing that the L22P gastric cancer-associated variant lacks dRP lyase and DNA-binding activities are consistent with the possibility that this variant is linked to human cancer. Cells sustain at least 20 000 oxidative lesions per cell per day and it is likely that cancer treatment induces additional DNA damage. Some of the oxidative base damage could be removed by a monofunctional DNA glycosylase. If this were the case, it is likely that in the presence of L22P BER is initiated, the gap is filled in, but ligation does not occur if dRP group remains in the DNA. Upon encountering a replication fork, the unligated nick could be converted to a double-strand break. If repaired improperly, end joining could lead to translocations or deletions. If these alterations occur in key growth control genes, it could lead to tumorigenesis or metastasis. However, much of the oxidative base damage is likely to be removed by bifunctional DNA glycosylases. When this occurs, it is unlikely that the lack of dRP Lyase activity of L22P plays a role in increasing genomic instability. Rather, the weak affinity of L22P for DNA could lead to many unfilled gaps in cells, which could be converted to double-strand breaks and lead to genomic instability. Alternatively, the gap could be enlarged by nucleases and polymerases other than pol β, such as Y family polymerases with dRP lyase activity that could fill in the gap in an error prone manner, resulting in mutations and leading to cancer. Preliminary studies in our laboratory, showing that expression of L22P in mouse fibroblasts induces cellular transformation, (Dalal and Sweasy, unpublished data), support this idea. The presence of the dRP lyase-deficient L22P is likely to be important during treatment of the tumor by drugs including alkylating agents. These agents result in cytoxicity in the absence of DNA repair, but it is likely that many of the lesions are repaired by the BER pathway. It is likely that interference with BER leads to increased genomic instability.
Interestingly, we have shown that four of the other pol β cancer-associated variants, namely, K289M, I260M (14), E295K (16) and Y265C (Sweasy, unpublished data), induce cellular transformation when expressed in mouse fibroblasts. Each of these variants is likely to induce cellular transformation by causing genomic instability. The K289M and I260M colon- and prostate cancer-associated variants are sequence-specific mutator polymerases (15,34). The gastric cancer-associated E295K variant is polymerase deficient and acts in a dominant negative manner to interfere with BER. This variant also induces sister chromatid exchanges in mouse cells (16). Our combined studies of these mutants support the mutator phenotype hypothesis of human cancer (35) and suggest that BER is a tumor suppressor mechanism.
ACKNOWLEDGEMENTS
We thank Nilesh Banavali for suggestions and helpful discussions on the evaluation of the molecular dynamics simulations. We also thank Reem Hanna for her assistance with the protein–protein interaction experiments. This research was supported by CA16038 awarded to J.B.S. from the National Cancer Institute. Funding to pay the Open Access publication charges for this article was provided by the National Cancer Institute. This research was also supported by Health Research Inc. New Investigator Funds (to J.J.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 2.Sung JS, Demple B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 2006;273:1620–1629. doi: 10.1111/j.1742-4658.2006.05192.x. [DOI] [PubMed] [Google Scholar]
- 3.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 4.Sobol RW, Horton JK, Kühn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 5.Sobol RW, Wilson SH. Mammalian DNA beta-polymerase in base excision repair of alkylation damage. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:57–74. doi: 10.1016/s0079-6603(01)68090-5. [DOI] [PubMed] [Google Scholar]
- 6.Fortini P, Parlanti E, Sidorkina OM, Laval J, Dogliotti E. The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J. Biol. Chem. 1999;274:15230–15236. doi: 10.1074/jbc.274.21.15230. [DOI] [PubMed] [Google Scholar]
- 7.Srivastava DK, Berg BJ, Prasad R, Molina JT, Beard WA, Tomkinson AE, Wilson SH. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J. Biol. Chem. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- 8.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd. Washington D.C: ASM Press; 1995. pp. 169–226. [Google Scholar]
- 9.Pascucci B, Maga G, Hubscher U, Bjoras M, Seeberg E, Hickson ID, Villani G, Giordano C, Cellai L, et al. Reconstitution of the base excision repair pathway for 7,8-dihydro-8-oxoguanine with purified human proteins. Nucleic Acids Res. 2002;30:2124–2130. doi: 10.1093/nar/30.10.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Maciejewski MW, Liu D, Prasad R, Wilson SH, Mullen GP. Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity. J. Mol. Biol. 2000;296:229–253. doi: 10.1006/jmbi.1999.3455. [DOI] [PubMed] [Google Scholar]
- 12.Prasad R, Batra VK, Yang XP, Krahn JM, Pedersen LC, Beard WA, Wilson SH. Structural insight into the DNA polymerase beta deoxyribose phosphate lyase mechanism. DNA Repair. 2005;4:1347–1357. doi: 10.1016/j.dnarep.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase beta and cancer? Cell Cycle. 2004;3:998–1001. [PubMed] [Google Scholar]
- 14.Sweasy JB, Lang T, Starcevic D, Sun KW, Lai CC, Dimaio D, Dalal S. Expression of DNA polymerase {beta} cancer-associated variants in mouse cells results in cellular transformation. Proc. Natl Acad. Sci. USA. 2005;102:14350–14355. doi: 10.1073/pnas.0505166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang T, Maitra M, Starcevic D, Li SX, Sweasy JB. A DNA polymerase beta mutant from colon cancer cells induces mutations. Proc. Natl Acad. Sci. USA. 2004;101:6074–6079. doi: 10.1073/pnas.0308571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lang T, Dalal S, Chikova A, Dimaio D, Sweasy JB. The E295K DNA polymerase beta gastric cancer-associated variant interferes with base excision repair and induces cellular transformation. Mol. Cell. Biol. 2007;27:5587–96. doi: 10.1128/MCB.01883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalal S, Kosa JL, Sweasy JB. The D246V mutant of DNA polymerase beta misincorporates nucleotides: evidence for a role for the flexible loop in DNA positioning within the active site. J. Biol. Chem. 2004;279:577–584. doi: 10.1074/jbc.M309607200. [DOI] [PubMed] [Google Scholar]
- 18.Haracska L, Prakash L, Prakash S. A mechanism for the exclusion of low-fidelity human Y-family DNA polymerases from base excision repair. Genes Dev. 2003;17:2777–2785. doi: 10.1101/gad.1146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobol RW. DNA polymerase beta null mouse embryonic fibroblasts harbor a homozygous null mutation in DNA polymerase iota. DNA Repair. 2007;6:3–7. doi: 10.1016/j.dnarep.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, et al. NAMD2: greater scalability for parallel molecular dynamics. J. Comp. Phys. 1999;151:283–312. [Google Scholar]
- 21.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.Humphrey W, Dalke A, Schulten K. VMD—visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 23.McKerell AD, Jr, Bashford D, Bellot M, Dunbrack R.L., Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. 1998;102:3583–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 25.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase Beta. Chem. Rev. 2006;106:361–382. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 26.Prasad R, Beard WA, Wilson SH. Studies of gapped DNA substrate binding by mammalian DNA polymerase beta. Dependence on 5′-phosphate group. J. Biol. Chem. 1994;269:18096–18101. [PubMed] [Google Scholar]
- 27.Chagovetz AM, Sweasy JB, Preston BD. Increased activity and fidelity of DNA polymerase beta on single-nucleotide gapped DNA. J. Biol. Chem. 1997;272:27501–27504. doi: 10.1074/jbc.272.44.27501. [DOI] [PubMed] [Google Scholar]
- 28.Lyu PC, Sherman JC, Chen A, Kallenbach NR. Alpha-helix stabilization by natural and unnatural amino acids with alkyl side chains. Proc. Natl Acad. Sci. USA. 1991;88:5317–5320. doi: 10.1073/pnas.88.12.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sobol RW, Watson DE, Nakamura J, Yakes FM, Hou E, Horton JK, Ladapo J, Van Houten B, Swenberg JA, et al. Mutations associated with base excision repair deficiency and methylation-induced genotoxic stress. Proc. Natl Acad. Sci. USA. 2002;99:6860–6865. doi: 10.1073/pnas.092662499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 31.Prasad R, Beard WA, Chyan JY, Maciejewski MW, Mullen GP, Wilson SH. Functional analysis of the amino-terminal 8-kDa domain of DNA polymerase beta as revealed by site-directed mutagenesis. DNA binding and 5′-deoxyribose phosphate lyase activities. J. Biol. Chem. 1998;273:11121–11126. doi: 10.1074/jbc.273.18.11121. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto Y, Kim K, Katz DS, Feng JA. Catalytic center of DNA polymerase beta for excision of deoxyribose phosphate groups. Biochemistry. 1998;37:6456–6464. doi: 10.1021/bi9727545. [DOI] [PubMed] [Google Scholar]
- 33.Feng JA, Crasto CJ, Matsumoto Y. Deoxyribose phosphate excision by the N-terminal domain of the polymerase beta: the mechanism revisited. Biochemistry. 1998;37:9605–9611. doi: 10.1021/bi9808619. [DOI] [PubMed] [Google Scholar]
- 34.Dalal S, Hile S, Eckert KA, Sun KW, Starcevic D, Sweasy JB. Prostate-cancer-associated I260M variant of DNA polymerase beta is a sequence-specific mutator. Biochemistry. 2005;44:15664–15673. doi: 10.1021/bi051179z. [DOI] [PubMed] [Google Scholar]
- 35.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]