Abstract
Microsatellites are ubiquitously present in eukaryotic genomes and are implicated as positive factors in evolution. At the nucleotide level, microsatellites undergo slippage events that alter allele length and base changes that interrupt the repetitive tract. We examined DNA polymerase errors within a [T]11 microsatellite using an in vitro assay that preferentially detects mutations other than unit changes. We observed that human DNA polymerase kappa (Pol κ) inserts dGMP and dCMP within the [T]11 mononucleotide repeat, producing an interrupted 12-bp allele. Polymerase β produced such interruptions at a lower frequency. These data demonstrate that DNA polymerases are capable of directly producing base interruptions within microsatellites. At the molecular level, expanded microsatellites have been implicated in DNA replication fork stalling. Using an in vitro primer extension assay, we observed sequence-specific synthesis termination by DNA polymerases within mononucleotides. Quantitatively, intense, polar pausing was observed for both pol κ and polymerase α-primase within a [T]11 allele. A mechanism is proposed in which pausing results from DNA bending within the duplex stem of the nascent DNA. Our data support the concept of a microsatellite life-cycle, and are consistent with the models in which DNA sequence or secondary structures contributes to non-uniform rates of replication fork progression.
INTRODUCTION
Approximately 3% of the human genome contains microsatellite DNA sequences, which are present on every chromosome at an average density of ∼14 000 bp/Mbp (1). Therefore, repetitive microsatellite DNA comprises a significant component of genome replication that must occur faithfully each cell cycle. Direct sequence analyses of eukaryotic genomes have revealed that the precise composition of microsatellites is heterogeneous, ranging from pure arrays of a single repetitive sequence, to complex arrays containing several types of repetitive units, to arrays that are interrupted by single base changes or insertion/deletion mutations (2). Sequence variation in common microsatellites has been proposed to have a positive role in evolution (3), and numerous reports have illustrated the phenotypic (gene expression) effects of microsatellite length variation (4,5). Evolutionary models of microsatellites propose that length distributions reflect a balance between expansion or contraction errors and point mutations within the allele (6). The accumulation of base interruptions has been proposed to break the repetitive array, resulting in death of the microsatellite (7). Expansion/contraction mutations are generally assumed to occur by slipped strand mispairing (8), and both in vivo and in vitro data are consistent with microsatellite errors that result in the gain or loss of repeat units (9–11).
Microsatellites also have been implicated in genome stability at the molecular level, through functional effects on chromatin organization, recombination and DNA replication (12). Expanded trinucleotide alleles can adopt non-B DNA secondary structures, thereby causing DNA polymerase pausing in vitro and replication fork pausing in vivo (13,14). Replication fork arrest may precede replication fork collapse, resulting in double strand breaks and genome rearrangements (15,16). In contrast to the trinucleotide microsatellites, little is known about the biochemistry of DNA replication through the more highly abundant, shorter, mono- and dinucleotide repetitive sequences. Like the trinucleotide repeats, these sequences also have the potential to adopt non-B form DNA structures (17). We previously observed strong DNA polymerase α-primase (pol α-primase) pauses within a [TC]20 dinucleotide allele that were caused by triplex DNA formation between the nascent duplex DNA and the ssDNA template during DNA synthesis (18).
The identification of DNA polymerases present at the human replication fork is currently under intense investigation (19,20). Current models to explain resolution of stalled replication forks invoke replacement of replicative polymerases or gap-filling behind the replication fork by specialized DNA polymerases (21–23). DNA polymerase κ (Pol κ) has been detected in nuclear foci at a low level in undamaged human cells (24), suggesting pol κ may be active at replication forks. Pol κ also is capable of performing efficient extension synthesis from mispaired and misaligned template-primer termini in vitro (25,26). Because human polymerases differ significantly in fidelity (27,28), the identity of the polymerase(s) synthesizing nascent microsatellite DNA is expected to impact microsatellite stability. In the current study, we investigated the biochemistry and accuracy of pol κ during DNA synthesis through mononucleotide alleles. Our mutagenesis studies show that DNA polymerases may influence the evolution of microsatellites by creating base interruptions, thereby rendering the microsatellite sequences more stable. Our DNA synthesis progression studies show that short microsatellites also may inhibit DNA polymerases by forming alternative DNA secondary structures.
MATERIALS AND METHODS
Reagents and vectors
Purified full-length (99 kDa) human pol κ was kindly supplied by Dr Zhigang Wang (University of Kentucky, Lexington, KY) or was purchased from Enzymax (Lexington, KY). Purified human pol α-primase complex was purchased from Chimerx (Madison, WI). The 3′→5′ exonuclease-deficient form of Escherichia coli DNA polymerase I large fragment (D424A) was a generous gift from Dr Catherine Joyce (Yale University). Recombinant DNA polymerase β was purified as described (29). The mononucleotide vectors containing [T/A]9 (inserted between bases 111 and 112 of the target HSV-tk gene) or [G/C]9 (inserted between bases 110 and 111) inserts were constructed by cloning products of an oligonucleotide-directed in vitro T7 DNA polymerase reaction as described (30). Bases flanking the inserts were included as part of the microsatellite region in data analyses when appropriate. Therefore, the total allele length of the microsatellite is always reported. The microsatellite unit located closest to the 5′ end of the HSV-tk sense strand was denoted as unit 1.
In vitro HSV-tk mutagenesis assay
Linear DNA fragments and ssDNA were prepared and used to construct gapped duplex (GD) molecules, as described (31), with the following modification. A StuI restriction site was created at HSV-tk position 180, and subcloned into the pSS vector (11). Restriction enzyme digestion with MluI and StuI and subsequent hybridization to ssDNA creates a hetereoduplex molecule containing an 81 nt gap that encompasses the microsatellite allele, but excludes the majority of the HSV-tk coding sequence. Oligonucleotide-primed ssDNA templates were constructed to initiate DNA synthesis at position 169 of the HSV-tk gene. The in vitro reactions contained 1 pmol of template DNA at 40 nM concentration. Reaction conditions for pol β were as described (11), except that 10 pmol enzyme were used. Reaction conditions for pol κ were as below, except that 8 pmol enzyme were used. To sample reaction products for mutations, small fragments were prepared by MluI and StuI restriction digestion and hybridized to the corresponding GD as described (11,31). Successful hybridization to GD was achieved for all reactions, as determined by agarose gel electrophoresis. To select for HSV-tk mutations, an aliquot of DNA from the final hybridization was used to transform recA13, upp, tdk E. coli strain FT334 by electroporation, and plated on VBA selective media (31). Bacteria harboring HSV-tk mutant plasmids were selected by plating in the presence of 40 µM 5-fluoro-2′-deoxyuridine and 50 µg/ml chloramphenicol. The DNA sequence of the HSV-tk gene in the MluI-StuI region of independent mutants was determined as described (11). Differences in proportions of specific types of mutations were analyzed statistically using Fisher's exact test (two-tailed).
Polymerase pausing
Pausing analyses were carried out as described (18). Briefly, DNA synthesis templates were created by hybridizing a [γ-32P] 5′-end labeled oligonucleotide to the appropriate ssDNA at a 1:1 molar ratio. The standard reactions for pol κ primer extension contained 25 mM potassium phosphate at the indicated pH, 5 mM MgCl2, 5 mM dithiothreitol, 100 μg/ml BSA, 1 mM dNTPs and 200 fmol primer-template. Synthesis was initiated upon addition of 50–100 fmol pol κ. Under these conditions, we routinely observe reaction products up to 100 nt or more in length, and the distribution of these products is independent of reaction time. Conditions for the pol α-primase reactions were as described (18). Analyses to test triplex potential were done analogously, substituting the nucleotide analog, 7-deaza-dATP, for dATP. Quantitation of reaction products was performed using a Molecular Dynamics Phosphorimager and ImageQuant software (Sunnyvale, CA). Total percent extension is the amount of total extended molecules (corrected for percent hybridization and background) divided by this number plus the amount of corrected primer molecules. The percent synthesis termination at a particular nucleotide was calculated by dividing the number of reaction product molecules of that length by the total number of template-primer molecules extended in the reaction.
RESULTS
Polymerase mutagenesis within a [T]11 mononucleotide allele
The sequence composition of microsatellites is non-random, and in the human genome, poly(dA/dT) mononucleotides are the most abundant class (1,32). In this study, we set out to determine mutational mechanisms within mononucleotide alleles using purified human DNA polymerases. We previously developed an in vitro assay to measure DNA polymerase errors within microsatellite alleles, using the HSV-tk gene as a mutational reporter (11). In the published system, polymerase errors are produced within either an artificial in-frame microsatellite sequence or the HSV-tk coding region, and are detected phenotypically using selective plating of E. coli. In our current study, we modified our existing assay by excluding ∼100 nt of target template sequence within the coding region in order to improve detection of polymerase errors within the microsatellite allele. Our first modified HSV-tk construct contained an in-frame [T]11 microsatellite (constructed by inserting a T9 sequence within an existing TT sequence of the HSV-tk gene). The major types of cellular mutational events at mononucleotide alleles are insertions or deletions of one repeat unit (33–35). However, in E. coli, out-of-frame mutations that occur in repetitive poly(dA) or poly(dT) sequences longer than 9 nt can be placed back in-frame by transcriptional slippage during RNA polymerase elongation (36). In agreement with this phenomenon, we have confirmed that out-of-frame [T]10 and [T]19 alleles do not produce an HSV-tk mutant phenotype, whereas an out-of-frame [T]7 mutant was detectable (data not shown). Thus, our modified mutational assay is biased against detection of one unit changes within our [T]11 allele, allowing improved detection of non-canonical mutations.
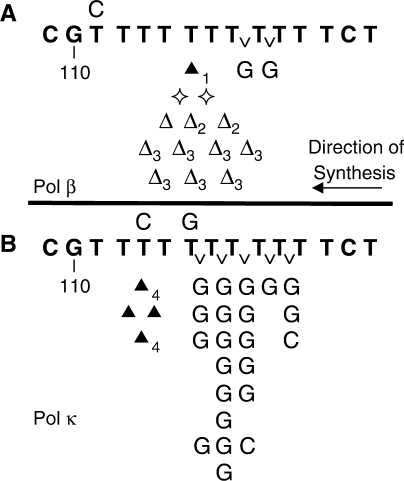
We used this specialized mutagenesis assay to determine the frequency and specificity of errors produced by human DNA polymerases κ and ß. For pol κ, we observed that the majority of HSV-tk mutants contained errors within the coding region (Table 1). Thirty percent of pol κ errors were found within the [T]11 microsatellite sequence. Of these, the majority (78%) of errors observed are insertion of a dGMP or dCMP residue within the [T]11 microsatellite sequence (Table 1), corresponding to an error frequency of 5.6 × 10–4. The pol κ insertions are not evenly distributed throughout the microsatellite allele, but primarily occur midway during DNA synthesis, between positions T6 and T7 or between T7 and T8 (Figure 1). These alleles produce new out-of-frame microsatellite loci (for example, T6GT5) that are detectable mutants. DNA synthesis by pol β produced a higher proportion of errors (40%) within the [T]11 microsatellite sequence, relative to pol κ. In contrast to pol κ, however, the majority of pol β mutants are either deletion of one T nucleotide within the microsatellite allele (detected as a multiple mutation) or complex errors involving a T deletion within the microsatellite (see legend to Figure 1). Only 12% of pol ß microsatellite errors are non-iterative base insertions, corresponding to a frequency of 1.8 × 10–4 (Table 1). Thus, pol κ and pol β show a statistically significant difference in the proportion of insertion errors within the microsatellite (P < 0.0001, Fisher's exact test). The mutational specificity difference between the polymerases is specific to the microsatellite region, as the two polymerases produce a similar proportion and type of frameshift errors within the coding region (P = 0.77, Fisher's exact test) (Table 1).
Table 1.
Mutational specificity of DNA polymerases κ and β
| Region/mutational classa | Number of events (Proportion of region) | |
|---|---|---|
| pol κ | pol β | |
| T11 microsatellite | 28 | 16 |
| T deletionb | 0 | 12 (0.75) |
| T insertionc | 4 (0.14) | 1 (0.06) |
| G insertion | 20 (0.71) | 2 (0.12) |
| C insertion | 2 (0.07) | 0 |
| Base substitutiond | 2 (0.07) | 1 (0.06) |
| HSV-tk coding | 65 | 24 |
| Base substitutions | 9 (0.14) | 3 (0.12) |
| Frameshiftse | 53 (0.82) | 19 (0.79) |
| Complex | 3 (0.05) | 2 (0.08) |
| HSV-tk− Mutant Frequencyf | 24 × 10−4 | 35 × 10−4 |
aMutations that create alleles longer than T9 do not result in a mutant phenotype due to transcriptional slippage within the microsatellite.
bIncludes −2T errors, −1T occurring as a multiple mutation, and complex errors involving the microsatellite.
cIncludes + 1T errors occurring as a multiple mutation and complex errors involving the microsatellite.
dBase substitution errors within the microsatellite occurred as multiple mutations.
eAll frameshifts were deletion errors for both polymerases.
fThe ssDNA mutant frequency is 1.3 × 10−4 for this template.
Figure 1.
Mutational specificity of DNA polymerases in the [T]11 assay. Middle sequence is the microsatellite target. Base substitutions are indicated above the sequence and frameshifts are indicated below. Symbols: open triangle: one T deletion (paired with a large 28bp coding region deletion); filled triangle: one T insertion (paired with a C to T at 100 or a C to T at 130); open diamond: two T deletion; letter ‘v’: insertion of noncanonical base. Subscripts indicate multiple errors detected within a single mutant. 1, T insertion paired with G to T at 110; 2, T deletion paired with T to C at STR; 3, T deletion paired with C 109 deletion; 4, T insertion paired with G insertion.
DNA synthesis termination within a [T]11 mononucleotide allele
In addition to nucleotide sequence changes that result in allelic polymorphism, microsatellite sequences can adopt non-B DNA forms that inhibit DNA synthesis. To examine whether DNA synthesis is inhibited within short mononucleotide alleles, we quantitated pol κ pausing within the [T]11 sequence using an in vitro primer extension assay under standard reaction conditions of 30°C and pH 7.0 (Figure 2). Termination profiles were determined empirically using enzyme to template ratios that limit polymerase rebinding to previously extended DNA products. For example, we observed 16% total primer-template extension after 30 min (lane 4) and this distribution is independent of time (lanes 2–5). Similar results were obtained for total percent extension values between 5 and 40%; therefore, comparisons of subsequent quantitative analyses within the 5′-[STR]-3′ templates were made using reactions with a percent extension within this range. For microsatellites within the complementary strand, we note that a higher total percent extension by pol κ is necessary to achieve enough synthesis through the microsatellite for quantitative analyses, as shown previously for pol α-primase (18).
Figure 2.
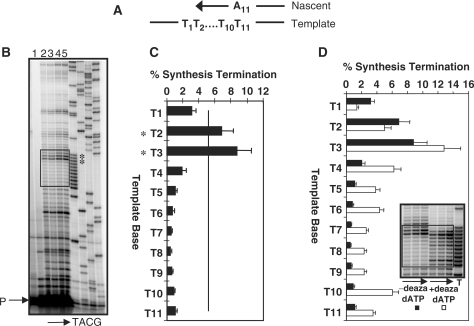
Polymerase κ termination within the T11 microsatellite. (A). Cartoon depicting DNA synthesis through the allele. (B). Representative phosphorimager scan of primer extension reaction products. Reactions were incubated at pH 7.0 and 30°C. Arrow indicates reaction time from 5 to 60 min. P, unextended primer. Lane 1, no polymerase control. Lanes 2–5, products of 5, 15, 30 and 60 min reactions. The 30 min reaction gave a total percent extension of 16.5%. Boxed region is the microsatellite. (C) Quantitation (30 min) for each template position in the microsatellite (mean of seven reactions). The continuous line represents the value of the percent synthesis termination that is 1 SD above the mean of all values within a given microsatellite. Asterisks indicate pause sites. (D) Deaza-dATP effect on termination. Quantitation of 30 min reactions without deaza-dATP (filled bars) or with deaza-dATP (open bars, three independent reactions). Inset: representative scan of primer extension products, with arrows indicating reaction time (5–30 min) and a box to indicate the beginning and end of the microsatellite sequence.
The percent synthesis termination was quantitated for each nucleotide within the [T]11 microsatellite (Figure 2C). The overall mean percent termination for the 11 nt within this microsatellite was 2.40 ± 0.43 (Table 2). A template position was considered a polymerase pause site if the percent synthesis termination for that residue was 1 SD above the mean of all microsatellite sites (shown as a continuous line in Figure 2C). We observed two prominent pol κ pauses at the 3′ end of the nascent strand, at residues T2 and T3. The sites of intense pausing (Figure 2) are not related to the sites of non-iterative base insertions (Figure 1). To examine whether the pronounced pausing within poly(dT) microsatellite sequences is polymerase-specific, we also examined pausing by the replicative polα-primase complex under its standard reaction conditions of 37°C and pH 7.5. A polar pausing pattern again was observed for pol α-primase synthesis. Pol α-primase displayed a mean percent termination per nucleotide of 4.78 ± 0.88, ∼4-fold greater than pol κ at a similar reaction temperature of 35°C (Table 2). Although reaction conditions were not identical, termination by polα-primase within residues T1 through T5 was 5–9-fold more intense than was pol κ (Figure 3C).
Table 2.
Polymerase κ pausing profile at mononucleotide microsatellite sequences
| STR sequence context | pH | Temperature (°C) | Mean percent termination per nta |
|---|---|---|---|
| 5′-[STR]-3′ | |||
| T11 | 6.5 | 30 | 2.39 (2) |
| 7.0 | 30 | 2.40 ± 0.43 (7) | |
| 7.0 + deaza-dAb | 30 | 4.57 ± 0.66 (3) | |
| 7.0 | 35 | 1.08 (2) | |
| 7.0 | 40 | 0.49 (2) | |
| G10 | 7.0 | 30 | 1.29 ± 0.07 (4) |
| 7.0 | 35 | 1.22 (2) | |
| 7.0 | 40 | 1.00 ± 0.13 (3) | |
| A11 | 7.0 | 30 | 1.28 ± 0.06 (5) |
| 3′-[STR]-5′ | |||
| C10C | 7.0 | 30 | 1.31 ± 0.14 (3) |
| A11C | 7.0 | 30 | 0.51 ± 0.15 (5) |
aData are the mean or the mean ± SD for the number of trials given in parentheses. Percent termination per nt is the amount of synthesis termination within the entire microsatellite, relative to total synthesis, normalized for the number of nucleotides in the allele.
bReactions contained 7-deaza-dATP in place of dATP.
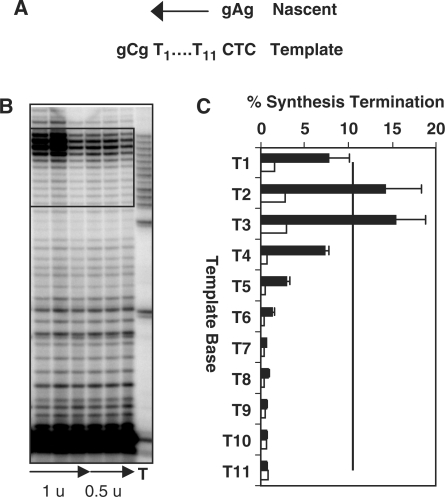
Figure 3.
Polymerase α-primase termination within the T11 microsatellite. (A) Cartoon depicting DNA synthesis through the allele. (B) Representative phosphorimager scan of primer extension reaction products. Pol α reactions contained either 0.5 or 1 unit of enzyme and were incubated at 37°C. Arrow indicates increase in reaction time from 5 to 30 min. (C) Quantitation of the percent synthesis termination (30 min) for each template position in the microsatellite for pol α-primase at 37°C (filled bars, three experiments) and pol κ at 35°C (open bars, two experiments).
Mechanism of polar pausing within the [T]11 microsatellite
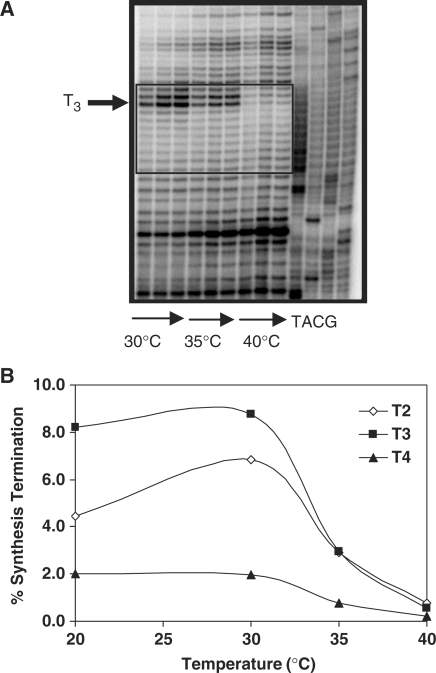
Poly(dA/dT) sequences have the potential to form intermolecular triplexes in vitro (37). We directly tested whether intramolecular triplex DNA structures were involved in polar pausing. The high pol κ termination probability within the [T]11 allele was not affected by lowering the reaction pH to 6.5 (Table 2), or by replacing dATP with 7-deaza-dATP (Figure 2D, Table 2). The enhanced termination observed upon addition of 7-deaza-dATP is consistent with published reports showing a decreased catalytic efficiency for incorporation of 7-deaza-dATP, relative to dATP, by pol κ (38). We observed strong, polar pauses at T1 and T2 by exonuclease-deficient Klenow polymerase; the pauses were somewhat reduced, but not eliminated, by addition of deaza-dATP (Supplementary data, Figure S1). We next examined whether the pol κ pause sites are temperature-dependent. We observed a 5-fold decrease in overall pol κ termination within the [T]11 allele as the reaction temperature was increased from 30°C to 40°C (Table 2), and the strong pause sites at positions T2 and T3 were eliminated at the higher reaction temperature (Figure 4A). Site-specific quantitation revealed that the percent synthesis termination decreased 9–15-fold as the temperature increased from 30°C to 40°C (Figure 4B).
Figure 4.
Effect of reaction temperature on polymerase κ termination at the T11 allele. (A) Representative phosphorimager scan of primer extension reaction products. Arrows indicate reaction time (5–30 min). Boxed area shows the beginning and end of the microsatellite sequence. (B) Variation of percent synthesis termination with temperature for nucleotides T2, T3 and T4. Each point represents mean of two independent experiments (30 min).
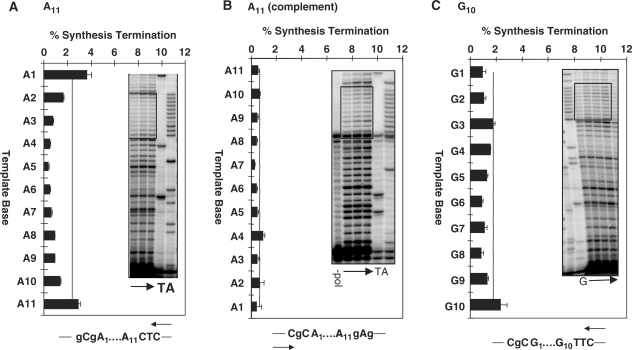
To determine whether the pronounced pausing within the [T]11 allele is sequence-dependent, we quantitated the pausing profiles along an [A]11 and a [G]10 mononucleotide allele within the same sequence context, and [A]11C and [C]10C alleles within the complementary sequence context (Figure 5). Data are graphed using the same scale as that for the T11 allele, in order to display the stark difference between pol κ termination at the [T]11 sequence versus the other mononucleotides. The mean percent termination per nucleotide within the [A]11 allele was 1.28 ± 0.06, while that within the [G]10 allele was 1.29 ± 0.07, both values approximately 2-fold lower than the [T]11 allele (Table 2). We also calculated the termination probability (39) within each allele in order to control for potential differences in the extent of polymerase termination within sequences prior to the microsatellites. Comparison of the termination probability for pol κ synthesis within the [T]11 versus the [A]11 allele again demonstrates that pol κ displays enhanced pausing within the poly(dT) sequence (Supplementary data, Figure S2). In addition, we examined the [A]11C allele in which the repetitive A tract is in the opposite sequence context, complementary to that of the [T]11 allele. In this case, termination at the [A]11C allele was very weak, 5-fold lower than that of [T]11 (Figure 5B, Table 2). In contrast to the complementary [T/A]11 pair, the mean percent termination per nucleotide was the same along the complementary [G]10 and [C]10C alleles (Table 2).
Figure 5.
Sequence dependence of pol κ termination within mononucleotide alleles. Phosphorimager quantitation of percent synthesis termination for 30 min reactions at pH 7.0 and 30°C are shown for (A). the A11 allele (mean of five experiments); (B) the A11 allele in the complementary sequence context (mean of five experiments); and (C) the G10 allele (mean of four experiments). Insets show representative phosphoimager scans of primer extension products, with arrows indicating reaction time (5–30 min). Boxed areas indicate the beginning and end of each microsatellite allele. Sequence below graph indicates sequence context of microsatellite and arrows indicate direction of synthesis. –pol, no polymerase control, showing that the dark band at the 3′ end of the A11C allele is an impurity in the primer and not a pause site.
DISCUSSION
Production of mononucleotide interruptions by DNA polymerases
We used a specialized microsatellite mutagenesis assay to determine the frequency and specificity of DNA pol κ and pol β errors within a [T]11 microsatellite. Strikingly, we observed that pol κ microsatellite errors involved the insertion of a dGMP or dCMP residue approximately midway within the [T]11 microsatellite sequence (Figure 1). These insertion events were specific for pol κ, as they were observed infrequently with pol β, and for the microsatellite, as pol κ did not produce such insertions within the downstream HSV-tk coding region (Table 1). These results demonstrate that DNA polymerases in vitro can directly create interrupted alleles from a pure microsatellite. Interrupted microsatellite alleles have been identified in phylogenetic genomic comparisons and may be responsible for microsatellite allele shortening (40). For example, a single base interruption of a poly(dT) locus was observed among primates lineages that produced a monomorphic, interrupted allele from a polymorphic, pure allele (41). In yeast, base interruptions of microsatellites result in increased genetic stability, relative to pure alleles (42). We suggest DNA synthesis by pol κ will promote mononucleotide stability because a major type of error it produces (interruptions) within the microsatellite are protective mechanistically.
DNA polymerase production of interrupted alleles occurred primarily through insertion of a non-iterative base within the repetitive microsatellite sequence, rather than by a base substitution mutation within the allele (Figure 1). Insertion of a non-iterated base within a repetitive sequence is extremely unusual in the literature of DNA polymerase fidelity, but has been occasionally detected by pol ζ (43), pol λ (44), pol η (45) and by pol ß using a damaged DNA template (46). Polymerase one-base insertion errors typically involve the insertion of the same nucleotide within a repetitive DNA sequence, or insertions of a base within a non-repetitive sequence (27). Several mechanisms for the in vitro production of misalignment-mediated DNA synthesis errors by purified DNA polymerases have been described (27,28). Deletions at non-iterative sequences have been proposed to be initiated by mispairing, followed by a template-primer misalignment. We propose that the pol κ dGMP insertions within the [T]11 microsatellite are initiated by formation of a T–dCMP mispair. Previous fidelity studies of pol κ have shown that the most frequently formed base substitution by pol κ is a T → G error, consistent with a high rate of T–dCMP mispairing (47,48). The highly repetitive nature of the [T]11 template sequence may facilitate rearrangement of the nascent DNA containing the dCMP to a misaligned intermediate by slippage elsewhere in the repetitive tract. In support of this model, we observe single T→G base substitutions within the microsatellite allele, which is the expected outcome in the absence of the misalignment step. An alternative model to explain the production of insertion mutations is transient misalignment (27), in which the primer strand relocates from position T6-T10 (the point of insertion) to position T1, with a concomitant template loop out of 4–8 bases. In this model, the dCMP insertion is templated by the G at position 110, followed by realignment of the primer strand. These two models can be differentiated experimentally by examining the effects of 5′ sequence context on the specificity of interruptions. Importantly, either model proposes that strand slippage within the repetitive microsatellite is involved in the production of interrupted alleles.
DNA structure and polymerase pausing
The study of replication dynamics through common microsatellite sequences and the key polymerases involved is crucial for understanding genome stability. We previously reported that the extent of DNA synthesis termination by the replicative, four subunit DNA pol α-primase complex within microsatellite sequences is unique for each microsatellite allele examined (18). Strong DNA pol α-primase pause sites within a [TC]20 allele were caused by triplex DNA formation between the nascent DNA primer-template duplex and the downstream template. We have observed strong, triplex-related pause sites for termination by pol κ within the same [TC]20 allele (data not shown). To explore the generality of DNA structure-induced polymerase pausing within common microsatellites, we examined termination within mononucleotide sequences. We observed intense, polar pause sites within a [T]11 allele during DNA synthesis by both pol κ and pol α-primase (Figures 2 and 3). However, we continued to observe the prominent [T]11 polar pause sites in the presence of 7-deaza-dATP (Figures 2 and S1). Therefore, the strong pause sites within the [T]11 allele do not result from intramolecular triplex DNA formation. We considered whether the conformation of the duplex primer stem formed during DNA synthesis through the microsatellite could affect polymerase termination. Poly(dA/dT) sequences are associated with altered DNA flexibility and the formation of bent DNA (49). A unique feature of bent DNA structure is that the tract displays a polarity, with the minor groove progressively narrowing from the 5′ end to the 3′ end of the track (50,51). DNA bending is also temperature dependent (52). Both of these features, polarity and temperature-dependence, were observed for the pol κ pause sites within the [T]11 template (Figures 2 and 4). Moreover, the cooperative unit for bending is ∼5 bp, and regions of uniform bent structure are observed in tracts longer than 7 bp (50,51). The major polymerase pause sites at T3 and T2 would correspond to a primer duplex stem length of eight and nine T–A basepairs, respectively. We propose that the observed pause sites within the [T]11 template result from structural changes to the duplex primer-template stem that disrupt polymerase–DNA interactions. Bent DNA displays a gradual shortening of the N3T-N1A hydrogen bond (51), and DNA polymerases utilize hydrogen bonding with minor groove acceptor sites to position the DNA primer stem within the active site for catalysis (53). Alternatively, bent DNA formed within the duplex primer stem may affect polymerase DNA affinity during synthesis. If the bent DNA model is correct, then we must also conjecture that the nucleotide composition of the nascent DNA strand directly affects the biochemical properties of the duplex stem, because we observed less pausing within an [A]11 allele in the same sequence context (Table 2). We note that along the [A]11 tract, a pause site is observed at position A1, at the very 3′ end of the allele. Possibly, the degree of duplex primer-stem bending is reduced or occurs with a longer phasing for the Atemplate–Tprimer sequence, relative to the Ttemplate–Aprimer sequence. Further experimentation is needed to fully test this model.
The rate of replication fork progression is not uniform through the eukaryotic genome (54,55), and replication fork stalling can be observed under non-stressed conditions (14). Previously, we proposed that the strong strand bias for pol α-primase termination within di- and tetranucleotide alleles may lead to a non-uniform rate of lagging strand DNA synthesis. The new results reported here for intense DNA polymerase pausing within poly(dT) sequences demonstrate that the inhibition of DNA synthesis within common microsatellites is sequence-specific. Thus, primary DNA sequence and accompanying DNA structural changes may be a factor contributing to nonrandom replication fork movements observed in vivo. In addition, the rate of replication fork progression through microsatellites will be controlled by the identity of the active polymerase, as we have shown that polα-primase termination at the [T]11 allele is much more intense than is pol κ termination (Figure 3C).
CONCLUSION
The human genome contains an abundance of poly (dA/dT). Unit-length changes in microsatellite alleles can impact gene expression, and microsatellites are under selective pressure during evolution. The primary mutational mechanism at microsatellite alleles has been shown to be DNA polymerase slippage. We report here a very unusual mutational spectrum of non-canonical base insertion errors produced in vitro by DNA polymerases, particularly pol κ, within a [T]11 allele. These errors effectively interrupt the microsatellite sequence, and may be a mechanism whereby microsatellites are stabilized in the genome. We have previously reported the production of both canonical (unit-length) and non-canonical errors within di- and tetranucleotide repeats during in vitro DNA synthesis by DNA pol β (11). The non-canonical errors observed include single base deletions or complex base substitution and deletions, all of which produced interrupted alleles from pure microsatellites. Our in vitro data for DNA polymerases demonstrate the highly dynamic nature of microsatellite mutagenesis, in that not only the length but also the homogeneity of the microsatellite is in continual flux. In support of the concept of a microsatellite life-cycle (7), we have observed directly the creation of polymorphic, common microsatellite alleles through expansion and contraction errors of unit length. Conversely, as reported here, we have observed the death of common alleles through base interruption errors that create shorter, interrupted alleles.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This study was funded by National Institutes of Health [CA100060] and the Jake Gittlen Cancer Research Foundation. We thank Dr Joann Sweasy, Dr Kataryna Makova, Yogeshwar Kelkar and Sandeep Shah for their critical reading of the manuscript and helpful comments. Funding to pay the Open Access publication charges for this article was provided by the Gittlen Cancer Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers GK, MacAvoy ES. Microsatellites: consensus and controvery. Comp. Biochem. Physiol. Part B. 2000;126:455–476. doi: 10.1016/s0305-0491(00)00233-9. [DOI] [PubMed] [Google Scholar]
- 3.Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Rothenburg S, Koch-Nolte F, Rich A, Haag F. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc. Natl Acad. Sci. USA. 2001;98:8985–8990. doi: 10.1073/pnas.121176998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rockman MV, Wray GA. Abundant raw material for cis-regulatory evolution in humans. Mol. Biol. Evol. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- 6.Kruglyak S, Durrett RT, Schug MD, Aquadro CF. Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc. Natl Acad. Sci. USA. 1998;95:10774–10778. doi: 10.1073/pnas.95.18.10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buschiazzo E, Gemmell NJ. The rise, fall and renaissance of microsatellites in eukaryotic genomes. Bioessays. 2006;28:1040–1050. doi: 10.1002/bies.20470. [DOI] [PubMed] [Google Scholar]
- 8.Levinson G, Gutman GA. Slipped strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 9.Sia EA, Jinks-Robertson S, Petes TD. Genetic control of micrsatellite stability. Mutat. Res. 1997;383:61–70. doi: 10.1016/s0921-8777(96)00046-8. [DOI] [PubMed] [Google Scholar]
- 10.Richards RI, Sutherland GR. Simple repeat DNA is not replicated simply. Nature Genet. 1994;6:114–116. doi: 10.1038/ng0294-114. [DOI] [PubMed] [Google Scholar]
- 11.Eckert KA, Mowery A, Hile SE. Misalignment-mediated DNA polymerase beta mutations: comparison of microsatellite and frameshift error rates using a forward mutation assay. Biochem. 2002;41:10490–10498. doi: 10.1021/bi025918c. [DOI] [PubMed] [Google Scholar]
- 12.Li YC, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol. Ecol. 2002;11:2453–2465. doi: 10.1046/j.1365-294x.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- 13.Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 14.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability. Mutat. Res. 2006;598:103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 16.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Sinden RR. DNA Structure and Function. New York: Academic Press; 1994. [Google Scholar]
- 18.Hile SE, Eckert KA. Positive correlation between DNA polymerase alpha-primase pausing and mutagenesis within polypyrimidine/polypurine microsatellite sequences. J. Mol. Biol. 2004;335:745–759. doi: 10.1016/j.jmb.2003.10.075. [DOI] [PubMed] [Google Scholar]
- 19.Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat. Res. 2006;166:693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- 20.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 21.Lehmann AR, Fuchs RP. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair. 2006;5:1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair. 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 24.Ogi T, Kannouche P, Lehmann AR. Localisation of human Y-family DNA polymerase kappa: relationship to PCNA foci. J. Cell. Sci. 2005;118:129–136. doi: 10.1242/jcs.01603. [DOI] [PubMed] [Google Scholar]
- 25.Washington MT, Johnson RE, Prakash L, Prakash S. Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc. Natl Acad. Sci. USA. 2002;99:1910–1914. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfe WT, Washington MT, Prakash L, Prakash S. Human DNA polymerase kappa uses template-primer misalignment as a novel means for extending mispaired termini and for generating single-base deletions. Genes Dev. 2003;17:2191–2199. doi: 10.1101/gad.1108603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Diaz M, Kunkel TA. Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem. Sci. 2006;31:206–214. doi: 10.1016/j.tibs.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Bebenek K, Kunkel TA. Streisinger revisited: DNA synthesis errors mediated by substrate misalignments. Cold Spring Harb. Symp. Quant. Biol. 2000;65:81–91. doi: 10.1101/sqb.2000.65.81. [DOI] [PubMed] [Google Scholar]
- 29.Opresko PL, Shiman R, Eckert KA. Hydrophobic interactions in the hinge domain of DNA polymerase ß are important but not sufficient for maintaining fidelity of DNA synthesis. Biochemistry. 2000;39:11399–11407. doi: 10.1021/bi000698t. [DOI] [PubMed] [Google Scholar]
- 30.Eckert KA, Yan G. Mutational analyses of dinucleotide and tetranucleotide microsatellites in Escherichia coli: influence of sequence on expansion mutagenesis. Nucleic Acids. Res. 2000;28:2831–2838. doi: 10.1093/nar/28.14.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckert KA, Hile SE, Vargo PL. Development and use of an in vitro HSV-tk forward mutation assay to study eukaryotic DNA polymerase processing of DNA alkyl lesions. Nucleic Acids. Res. 1997;25:1450–1457. doi: 10.1093/nar/25.7.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toth G, Gaspari Z, Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacob KD, Eckert KA. Escherichia coli DNA polymerase IV contributes to spontaneous mutagenesis at coding sequences but not microsatellite alleles. Mutat. Res. 2007;619:93–103. doi: 10.1016/j.mrfmmm.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran HT, Keen JD, Kricker M, Resnick MA, Gordenin DA. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell. Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sia EA, Kokoska RJ, Dominska M, Greenwell P, Petes TD. Microsatellite instability in yeast: dependence on repeat unit size and mismatch repair genes. Mol. Cell. Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner LA, Weiss RB, Driscoll R, Dunn DS, Gesteland RF. Transcriptional slippage occurs during elongation at runs of adenine or thymine in Escherichia coli. Nucleic Acids Res. 1990;18:3529–3535. doi: 10.1093/nar/18.12.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sen A, Graslund A. Structural constraints regulating triple helix formation by A-tracts. Biophys. Chem. 2000;88:69–80. doi: 10.1016/s0301-4622(00)00200-3. [DOI] [PubMed] [Google Scholar]
- 38.Johnson RE, Prakash L, Prakash S. Biochemical evidence for the requirement of Hoogsteen base pairing for replication by human DNA polymerase iota. Proc. Natl Acad. Sci. USA. 2005;102:10466–10471. doi: 10.1073/pnas.0503859102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eckert KA, Kunkel TA. Fidelity of DNA synthesis catalyzed by human DNA polymerase alpha and HIV-1 reverse transcriptase: Effect of reaction pH. Nucleic Acids Res. 1993;21:5212–5220. doi: 10.1093/nar/21.22.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor JS, Durkin JM, Breden F. The death of a microsatellite: a phylogenetic perspective on microsatellite interruptions. Mol. Biol. Evol. 1999;16:567–572. doi: 10.1093/oxfordjournals.molbev.a026138. [DOI] [PubMed] [Google Scholar]
- 41.Blanquer-Maumont A, Crouau-Roy B. Polymorphism, monomorphism, and sequences in conserved microsatellites in primate species. J. Mol. Evol. 1995;41:492–497. doi: 10.1007/BF00160321. [DOI] [PubMed] [Google Scholar]
- 42.Petes TD, Greenwell PW, Dominska M. Stabilization of microsatellite sequences by variant repeats in the yeast Saccharomyces cerevisiae. Genetics. 1997;146:491–498. doi: 10.1093/genetics/146.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong X, Garg P, Stith CM, McElhinny SA, Kissling GE, Burgers PM, Kunkel TA. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006;34:4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase lambda. Implications for function. J. Biol. Chem. 2003;278:34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 45.Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase eta. J. Mol. Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 46.Eckert KA, Hile SE. Alkylation-induced frameshift mutagenesis during in vitro DNA synthesis by DNA polymerases a and ß. Mutat. Res. 1998;422:255–269. doi: 10.1016/s0027-5107(98)00206-1. [DOI] [PubMed] [Google Scholar]
- 47.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DinB1 gene. J. Biol. Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Yuan F, Xin H, Wu X, Rajpal DK, Yang D, Wang Z. Human DNA polymerase kappa synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Res. 2000;28:4147–4156. doi: 10.1093/nar/28.21.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neidle S. Oxford Handbook of Nucelic Acid Structure. New York: Oxford University Press; 1999. [Google Scholar]
- 50.Crothers DM, Haran TE, Nadeau JG. Intrinsically bent DNA. J. Biol. Chem. 1990;265:7093–7096. [PubMed] [Google Scholar]
- 51.Nadeau JG, Crothers DM. Structural basis for DNA bending. Proc. Natl Acad. Sci. USA. 1989;86:2622–2626. doi: 10.1073/pnas.86.8.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koo HS, Wu HM, Crothers DM. DNA bending at adenine. thymine tracts. Nature. 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 53.Kiefer JR, Mao C, Braman JC, Beese LS. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 54.Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- 55.Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.